Spira-Lenburg Lab
Mission Statement
Our focus is on translational research to better understand lung biology and disease using post-genomic technologies and computational tools. The long terms goals of our lab are two fold. On the one hand, we seek to leverage these approaches to improve the diagnosis, treatment, and prevention of lung disease. On the other hand, we seek to develop and apply new research approaches and to train physician-scientists and graduate students who can apply these tools in the setting of translational research.
Background
The sequencing of the human genome, together with the technological advances that enabled this accomplishment, have ushered in a new era of biomedical research and profoundly altered the culture of science. We now have the ability to profile the properties of a biological sample in incredible detail, and this has led to an explosion of data from which we can now glean important new aspects of normal and disease biology: leading to new strategies for disease treatment and prevention; as well as new strategies for disease diagnosis, risk assessment and personalized medicine. The post-genomic era has not only shifted the research paradigm from studies of single genes or pathways to large-scale studies that combine data mining of genome-wide datasets for hypothesis generation followed by experimental work to validate these hypotheses , but has also highlighted a key role for bioinformatics: the application of techniques from computer science and statistics to identify and make sense of interesting patterns in the ever-more complex datasets produced by genome-wide profiling technologies.
Our lab consists of molecular biologists, clinicians, and computational scientists working together to improve the diagnosis, treatment, and prevention of lung disease. Our various research programs are united by a variety of themes. One unifying theme is our use of comprehensive genome-wide gene-expression profiling (“transcriptome profiling”) to discover distinctions between disease states that provide us not only with clues as to how disease develops, but also provides sensitive and specific tools for disease detection. Second, much of our research utilizes the concept of an “airway-wide field of injury” that results from toxic exposures such as cigarette smoking, or lung diseases such as lung cancer. The field of injury concept very simply posits that these types of abnormalities affect the entire respiratory tract and that responses to these insults can be detected in readily collected samples from the bronchus, nose or mouth (Figure 1). Our demonstrated ability to measure disease processes in these types of samples not only makes it possible to study responses to disease and environmental insults in large number of individuals, but also raises the possibility of being able to diagnose disease, establish disease risk, or predict response to therapy using biomarkers measured in these types of specimens.
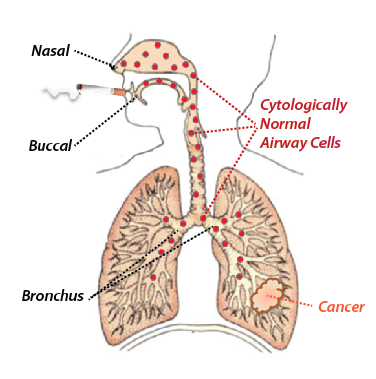 Figure 1: Concept of the “Field of Injury”
Figure 1: Concept of the “Field of Injury”
Selected Ongoing Research Projects
Using the field of injury our research focuses on:
The physiologic response to tobacco smoke. As many of our research goals are aimed at improving the treatment of patients with smoking-related lung diseases, we are interested in understanding how the body responds to tobacco smoke, and using this to better understand how tobacco smoke exposure contributes to disease. Using genomic approaches, we have identified smoking-related gene expression changes that occur throughout the respiratory tract and identified a subset that remain altered in people who have quit smoking. These irreversibly changed genes are especially interesting since disease risk remains elevated after smoking cessation. That many of the gene expression changes deep in the airway are also altered in cells that line the nose and mouth has led us to explore whether we can combine a simple upper airway test together with genome-wide approaches to answer basic questions such as: how physiologic responses to tobacco smoke vary amongst people who are exposed to different levels of tobacco smoke (or people who are only exposed to second-hand smoke), if differences in responses between individuals might contribute to differing levels of disease susceptibility, and if other inhaled pollutants cause similar differences in gene expression. This work is supported by grants from the NIEHS.
Detecting lung cancer. Using genomic approaches that allow us to comprehensively identify gene expression differences, we have identified a number of differences between smokers who have lung cancer and others who were thought to potentially have lung cancer but turn out instead to have a benign disease. We detect these expression differences in normal-looking cells from the large airway that are collected during bronchoscopy: a routine clinical procedure that is often employed as an early step in figuring out whether someone has lung cancer. We have shown that we can use a combination of several such genes as a biomarker that is both sensitive and specific for distinguishing smokers with lung cancer from those with benign disease and that this biomarker is more sensitive than the standard workup done as part of the routine bronchoscopy procedure. This biomarker has been licensed to an molecular diagnostic company (Allegro Diagnostics) that is seeking to validate its performance and make it available for clinicians to use as an adjunct to bronchoscopy. We are now determining whether these cancer-specific signals can also be detected in samples from the nose and whether there are gene expression differences that occur prior to the development of clinically detectable cancer in the hope that such differences could be used as a biomarker for assessing lung cancer risk. Lung cancer risk assessment could be used to determine which current and former smokers might benefit from increased lung cancer screening, or those who are good candidates for drugs that might prevent lung cancer. We are also exploring how this airway “field of injury” reflects alterations in oncogenic pathways that can be used to guide personalized genomic approaches to chemoprevention in high-risk smokers. This work is supported by grants from the NCI/EDRN and the Department of Defense.
Molecular classification and biomarkers for COPD and emphysema. Chronic Obstructive Pulmonary Disease (COPD) and emphysema are debilitating smoking-related lung diseases that often develop over an extended period and can have a remarkably different presentation and course between different patients. Using very similar approaches as our work in smoking and lung cancer, we have begun to identify gene expression differences that occur in the airway in patients with COPD and emphysema. Interestingly, the COPD-related gene-expression differences in the larger airways that we’ve studied are similar to the differences that occur in the small airways and alveolae: the tissues that are thought to be the main sites of disease. Moreover, these gene expression differences are more severe in patients with more severe disease and are diminished following treatment with anti-COPD therapy. These studies open the possibility of being able to molecularly dissect the clinical differences between patients with COPD using airway tissue readily obtained during bronchoscopy, and to develop biomarkers for monitoring a patient’s response to therapy. This work is supported by grants from the NHLBI.
Mechanisms of lung disease pathogenesis. In addition to developing biomarkers for assessing lung disease in clinical samples, we are also interested in using genome-wide approaches to improve our molecular understanding of lung disease pathogenesis. One strategy that we have used to model disease progression in both emphysema and lung cancer is to perform gene-expression profiling on multiple tissues from the same patient collected from regions of differing disease severity. Using this approach we have identified specific molecular processes involved in tissue remodeling that are specifically altered in regions of more severe emphysema. By combining these data with computational approaches to search databases of drugs, we have identified an existing drug as a potential emphysema therapeutic and validated that this drug reverses aspects of the emphysema gene expression signature and molecular defects in tissue remodeling pathways. We are also using this approach to explore the early molecular alterations that occur with progression of lung premalignancy.
A second approach has involved identifying microRNA expression differences associated with disease. While we are exploring using microRNA (miRNA) expression differences as the basis for biomarkers similar to our gene (mRNA) expression biomarkers, the regulatory function of miRNA makes them especially attractive for understanding the regulation of disease processes. The majority of our miRNA profiling work has been performed using high throughput RNA sequencing technology that has allowed us to develop a comprehensive portrait of all the small RNA that are expressed both in airway and lung tissue and discover a number of new miRNA. We have identified specific miRNA that are important regulators of the response to smoking as well as other miRNA that contribute to airway epithelial cell differentiation and repress aspects of lung carcinogenesis.
A third approach to understanding disease pathogenesis has involved the use of high throughput RNA sequencing to provide a comprehensive genome-wide view of the lung transcriptome at single nucleotide resolution. We are mining these data to identify disease-associated differences in transcript structure, expression of non-coding RNAs, etc. in the hope that they could serve as biomarkers, but more importantly that they might also provide specific clues as to the regulation of processes that contribute to disease pathogenesis.
Our work on the molecular regulation of the response to smoking and lung disease pathogenesis is supported by grants from the NIEHS, NHLBI and the Department of Defense.
Developing a Molecular Data Repository for the Lung Disease Research Community. This project, funded through The National Heart, Lung and Blood Institute (NHLBI), will establish the Lung Genomics Research Consortium (LGRC), a collaboration between Drs. David Schwartz (National Jewish Health), Naftali Kaminski (University of Pittsburgh), John Quackenbush (Dana-Farber cancer Institute), Marc Geraci (University of Colorado), Frank Sciurba (University of Pittsburgh), Ivana Yang (National Jewish Health) and Avrum Spira (Boston University). The goals of this project are to combine genetic, genomic and clinical information on greater than 400 human lung tissue samples from patients with various lung diseases (i.e. Interstitial Lung Disease (ILD), Chronic Obstructive Pulmonary Disease (COPD), & Emphysema).
Expertise
Gene-expression profiling
Transcriptome profiling via RNA-seq (next-generation sequencing using Illumina HiSeq technology)
Bioinformatics
Functional Genomics
Faculty
Avrum Spira, MD, MSc; Computational Biomedicine
Marc Lenburg, PhD; Computational Biomedicine
Gang Liu, PhD; Computational Biomedicine
Jennifer Beane, PhD; Computational Biomedicine
Adam Gower, PhD; Computational Biomedicine
Joshua Campbell, PhD; Computational Biomedicine
Vijaya Kolachalama, PhD, Computational Biomedicine
Ehab Billatos, MD; The Pulmonary Center, Computational Biomedicine
Sarah Mazzilli, PhD; Computational Biomedicine
Affiliated Faculty
Jerome Brody, MD; The Pulmonary Center
Frank Schembri, MD; The Pulmonary Center
Paola Sebastiani, PhD; School of Public Health
Daniel Brooks, MPH, DSc; School of Public Health
George O’Connor, MD, MS; The Pulmonary Center
Yuriy Alekseyev, PhD; Microarray Resource Center
Michael Platt, MD; Otolaryngology-Head & Neck Surgery|
Kei Suzuki, MD; Thoracic Surgeon, Boston Medical Center
Katrina Steiling, MD, MSc; The Pulmonary Center
Graduate Students
Kelley Anderson; Bioinformatics Program
Minyi Lee; Bioinformatics Program
Conor Shea; MD/Bioinformatics Program
Whitney Souery; MD/Bioinformatics Program
Dylan Steiner; Genetics and Genomics
Staff
Denise Fine: Clinical Research Coordinator
Erin Kane, PhD: Scientific Program Manager
Zhihan Li: Research Technician
Hanqiao Liu: Senior Research Technician
Shannon McDermott: Research Study Assistant
Austin Potter: Clinical Biospecimen Coordinator
Sherry (Xiaohui) Zhang: Senior Research Technician
Past Trainees
Beth Becker, PhD; Bioinformatics Program
Sean Corbett, PhD; Bioinformatics Program
Grant Duclos; Molecular & Translational Medicine Program
Jake Kantrowitz; Molecular & Translational Medicine MD/PhD Program
Julian Lel, MD; The Pulmonary Center
Boting Ning, PhD; Bioinformatics Program
Matt Nitzberg, MD; The Pulmonary Center
Ana Brandusa Pavel; Bioinformatics Program
Joseph Perez-Rogers; Bioinformatics Program
Gregory Radin, MD; The Pulmonary Center
Xingyi Shi, PhD; Bioinformatics Program
Anna Tassinari, PhD; Computational Biomedicine
Ke Xu, PhD; MD/Bioinformatics Program
Jiarui Zhang, PhD; Molecular & Translational Medicine Program
Collaborators (outside of BU medical center)
James Collins, PhD; College of Engineering, BU
Steven Chillirud, PhD; Columbia University
David Schwartz, MD; National Jewish Health
Ivana Yang, PhD; National Jewish Health
Naftali Kaminski, MD; University of Pittsburgh
Frank Sciurba, MD; University of Pittsburgh
Matthew Meyerson, MD, PhD; Dana-Farber Cancer Institute
John Quackenbush, PhD; Dana-Farber Cancer Institute
Mark Geraci, MD; University of Colorado
James Hogg, MD; University of British Colombia
Stephen Lam, MD; University of British Colombia
Andrea Bild, PhD; University of Utah
Pierre Massion, MD; Vanderbilt University
Steven Dubinett, MD; University of California, Los Angles
David Elashoff, PhD; University of California, Los Angles
Denise Aberle, MD; University of California, Los Angles
Steven Belinsky, PhD; Lovelace Respiratory Research Institute
Dirkje Postma, MD, PhD; Groningen University, Netherlands
Maarten van den Berge, MD; Gronigen University, Netherlands
Phil Hansbro, PhD, University of Newcastle, Australia
Selected Publications
- Wang, T.W., R.C.H. Vermeulen, W. Hu, G. Liu, X.J. Xiao, Y. Alekseyev, J. Xu, B. Reiss, K. Steiling, G.S. Downward, D.T. Silverman, F.S. Wei, G.P. Wu, J.H. Li, M.E. Lenburg, N. Rothman, A. Spira, Q. Lan. 2015. Gene-expression profiling of buccal epithelium among nonsmoking women exposed to household air pollution from smoky coal. Carcinogenesis. 36:1494-1501. PMID: 26468118.
- Silvestri, G.A., A. Vachani, D. Whitney, M. Elashoff, K. Porta Smith, J.S. Ferguson, E. Parsons, N. Mitra, J. Brody, M.E. Lenburg, and A. Spira, for the AEGIS Study Team. 2015. A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. New England Journal of Medicine. 373:243-251. PMID: 25981554.
- Campbell, JD, L Luo, G Liu, J Xiao, J Gerrein, BJ Guardela, J Tedrow, YO Aleksyev4, IV Yang, M Correll, M Geraci, J Quackenbush, F Sciurba, DA Schwartz, N Kaminski, S Monti, J Beane, A Spira, ME Lenburg. 2015. Assessment of microRNA differential expression and detection in multiplexed small RNA sequencing data. RNA. 21:164-71. PMID: 25519487
- Ooi AT, Gower AC, Zhang KX, Vick JL, Hong LS, Nagao B, Wallace WD, Elashoff D, Walser TC, Dubinett SM, Pellegrini M, Lenburg ME, Spira AE*, Gomperts BN*. 2014. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prevention Research. 7(5):487-95. (*co-senior authors)
- Perdomo, C., J.D. Campbell, J. Gerrein, C. Tellez, A. Gower, J. Vick, C. Garrison, C. Anderlind, G.R. Jackson, F. Schembri, B.N. Gomperts, P. Hayden, S.A. Belinsky, M.E. Lenburg*, A. Spira*. 2013. Identification of MiR-4423 as a Primate-Specific MicroRNA Highly Expressed in Airway Epithelium and Associated with Lung Cancer. Proceedings of the National Academy of Sciences. 110:18946-18951. (* contributed equally)
- Steiling, K., M. van den Berge, K. Hijazi, R. Florido, J. Campbell, G. Liu, J. Xiao, X. Zhang, Y. Alekseyev, D. Sin, P. Pare, J.C. Hogg, A. McWilliams, P.S. Hiemstra, P.J. Sterk, W. Timens, P. Sebastiani, G. O’Connor, D.S. Postma, S. Lam, A. Spira*, M.E. Lenburg*. 2013. Bronchial airway gene expression reflects a COPD-associated field of injury that changes with disease severity and is responsive to therapy. American Journal of Respiratory and Critical Care Medicine. 187:933-42. (* contributed equally)
- Campbell. J.D., J.E. McDonough, J.E. Zeskind, D.V. Pechkovsky, C.-A. Brandsma, M. Suzuki, J.V. Gosselink, G. Liu, Y.O. Alekseyev, J. Xiao, X. Zhang, S. Hayashi, J.D. Cooper, W. Timens, D.S. Postma, D.A. Knight, M.E. Lenburg*, J.C. Hogg*, A. Spira*. A gene expression signature of emphysema-related lung destruction and its reversal by the tripeptide GHK. Genome Medicine. 4(8):67, Aug 2012. * contributed equally
- Beane J, Cheng L, Soldi R, Zhang X, Liu G, Anderlind C, Lenburg ME, Spira A*, Bild A*. SIRT1 pathway dysregulation in the smoke-exposed airway epithelium and lung tumor tissue. Cancer Res. Sept 2012. * contributed equally
- Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO, Wang S, Levy S, Massion PP, Lenburg M, Spira A. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila). 6: 803-17, 2011.
- Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, Cooper K, Walker D, McWilliams A, Liu G, Szabo E, Brody J, Massion PP, Lenburg ME, Lam S, Bild AH, Spira A. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2(26): 26ra25, 2010.
- Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, Langer EM, Alekseyev Y, O’Connor GT, Brooks DR, Lenburg ME*, Spira A*. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 41(1): 1-8, 2010. * contributed equally
- Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Zhang X, Bowers J, Vaziri C, Ott K, Sensinger K, Collins JJ, Brody JS, Getts R, Lenburg ME, Spira A. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci 106(7): 2319-24. 2009.
- Beane J, Sebastiani P, Liu G, Brody J, Lenburg M, Spira A. Reversible and Permanent Effects of Tobacco Smoke Exposure on Airway Epithelial Gene Expression. Genome Biology. 8: R201, 2007.
- Spira A, Beane J, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas Y, Calner P, Sebastiani P, Sridhar S, Beamis J, Lamb C, Keane J, Lenburg M, Brody J. Airway Epithelial Gene Expression in the Diagnostic Evaluation of Smokers with Suspect Lung Cancer. Nature Medicine. 13:361-6. 2007.
- Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody J. Effects of Cigarette Smoke on the Human Airway Epithelial Cell Transcriptome. Proc Natl Acad Sci USA. 101:10143-8, 2004.