New Software Tool Aims to Help Scientists Better Analyze Complex Spatial Data From Tissues
Photo by Javier Miranda on Unsplash.
Research
New Software Tool Aims to Help Scientists Better Analyze Complex Spatial Data From Tissues
“Giotto Suite” to streamline the study of tissue biology and disease mechanisms
Researchers at Boston University Chobanian School of Medicine, Icahn School of Medicine at Mount Sinai in New York and Boston Medical Center have developed a software platform to help scientists more easily analyze the molecular structure of tissue in both healthy and disease states. Details on the platform, called Giotto Suite, were reported in the Oct. 1 online issue of Nature Methods.
In recent years, new technologies have made it possible to capture detailed maps of RNA and proteins within intact tissues—a field known as spatial omics. These advances are providing valuable insight into how cells behave and interact in different tissue microenvironments, which is especially important for understanding conditions such as cancer, neurodegenerative diseases, and immune disorders.
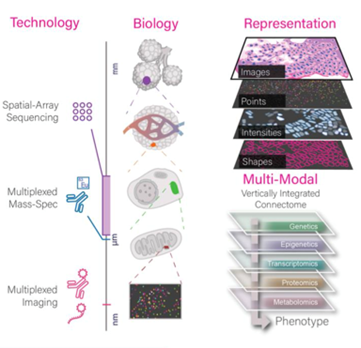 Illustration depicting the conceptual and flexible integration of spatial datasets across multiple length scales and data modalities in Giotto Suite. Chen, Chávez-Fuentes, et al., Nature Methods
Illustration depicting the conceptual and flexible integration of spatial datasets across multiple length scales and data modalities in Giotto Suite. Chen, Chávez-Fuentes, et al., Nature Methods
“While spatial omics technologies are generating incredibly rich data about the molecular landscape of tissues, the real challenge now lies in analyzing and interpreting that data,” says co-senior corresponding author Guo-Cheng Yuan, PhD, professor of genetics and genomic sciences, at Icahn School of Medicine at Mount Sinai. “The biggest obstacle is no longer generating the information—it’s making sense of it. Biologists need tools that can keep pace with the growing complexity and scale of these datasets and help them draw meaningful conclusions. We wanted to create a tool that not only simplifies the analysis process, but also adapts to different types of spatial data and workflows, so researchers can focus on the science rather than the technical hurdles.”
To meet this need, the research teams created Giotto Suite, a suite of modular software tools built in R, a widely used programming language for data analysis. The tool is designed to work with spatial data from any platform and supports a broad range of analyses, including data integration across different types of measurements and resolutions. Its flexible structure also allows users to begin analysis from multiple points, depending on their research needs, say the investigators.
 Ruben Dries, PhD
Ruben Dries, PhD“Many existing tools require users to combine multiple packages to complete a full analysis,” says co-senior corresponding author Ruben Dries, PhD, assistant professor of medicine, hematology and medical oncology at BU Chobanian & Avedisian School of Medicine and associate member of the Center for Regenerative Medicine (CReM). “This tool brings those steps into a single framework, aiming to streamline the process. We also developed it with scalability in mind to accommodate the growing size and complexity of spatial datasets. Our goal was to develop a platform that helps other researchers uncover new insights from their spatial data.”
The researchers cautioned that the current version includes only a subset of available analytical methods, which may not suit every research question. They emphasized, however, that the platform is intended to evolve and that they plan to continue expanding its features, enhancing its interoperability with external software packages, and offering training and support to the research community.
Looking ahead, the researchers plan to apply Giotto Suite in their own studies of cancer and neurodegenerative diseases, while continuing to refine the tool based on feedback from the scientific community. The tool is freely available to researchers at http://giottosuite.com, and the team hopes it will encourage wider use of spatial omics technologies in biomedical research.