Boston University Microarray Resource
Overview

The Boston University Microarray Resource provides in-house analysis of global gene expression and is available to all researchers at Boston University. We are located on the 6th floor of the “E” building at the BU Medical Campus.
Currently, we provide analysis of gene expression using the Affymetrix GeneChip platform. Services using the Illumina platform will be available soon. For more information go to the services section of our website. For both platforms users of the facility provide us with high quality samples and the Microarray Resource does the rest. We also offer analysis of RNA, DNA and Proteins using Agilent Bioanalyzer.
We offer the same services to researchers outside of Boston University on an available-resource basis.
All interested researchers are encouraged to contact the Microarray Resource to discuss potential projects or to ask any question about the use of microarrays. Dr. Alekseyev can provide advice on experimental design, RNA isolation methods, and just about any aspect of array hybridization. Dr. Lenburg can provide advice on experimental design and data analysis.
We look forward to talking with you!
People

Yuriy Alekseyev, Ph.D.
Research Assistant Professor
Director, Microarray Core Facility
E605
(617)414-1377
yurik@bu.edu

Marc Lenburg, Ph.D.
Associate Professor
co-Director, Microarray Core Facility
E613B
(617)414-1375
mlenburg@bu.edu
Introduction to Microarrays
Biological arrays are nothing more than an ordered set of compounds affixed to a solid surface. By applying a sample solution to the array it is possible to assay the interaction of the sample with each of the compounds on the array. Microarrays are a powerful research tool because they enable massively-parallel assays of biological samples.
The most common application of microarrays is gene expression analysis. In this case the interaction between labeled mRNA in a biological sample and complementary DNA probes, which are affixed to the array, allows rapid quantification of the entire transcriptome. Many other types of arrays are possible. Nucleic acid arrays can also be used for genotyping and antibody arrays can be used for proteomics.
The Affymetrix GeneChip system is a commercial platform that enables rapid, reproducible, and accurate microarray analysis on a genome-wide scale. The Affymetrix GeneChip platform can be used for gene expression, chromatin immunoprecipitation and genotyping analysis: really anything that involves sequence-specific nucleic-acid hybridization.At the Boston University Microarray Resource we strive to allow you to easily incorporate microarrays into your research and help you get the most out of your data. You provide us with high quality samples and the Microarray Resource does the rest.servicesAffymetrix GeneChip® System for Gene Expression AnalysisThe Affymetrix GeneChip system is a commercial microarray platform that allows whole genome gene expression analysis for a wide variety of experimental organisms (http://www.affymetrix.com/products/arrays/index.affx). This system has three major advantages over other array systems: it is easy to get rapid results; it has the capability to monitor the expression of every gene in the genome; and it is the most widely used commercial microarray platform.
Getting started with Affymetrix GeneChips is easy:
- Set-up an appointment with members of the Microarray Resource to discuss your experiment. This is optional, but highly recommended.
- Give us total RNA (Standard processing is available starting with as little as 50 ng of total RNA. Exon arrays require 1 µg of total RNA)
- Wait for us to process your samples
- Work with Microarray Resource to analyze data
Affymetrix GeneChip® Gene 1.0 ST Array System for Human, Mouse and RatThe Affymetrix GeneChip Gene 1.0 ST Array System represents a new generation of array products and offers a number of important advantages: it is more cost-effective for gene expression profiling, and measures expression using probes that span the full length of the transcript (unlike previous Affymetrix arrays that used probes targeted to the 3’ end of the transcript).Affymetrix GeneChip® System for Exon-level Gene Expression AnalysisThe Affymetrix GeneChip Exon Array system provides, for the first time, exon-level expression profiling of the whole-genome on a single array. The arrays contain probes for over 1 million exon clusters within regions of the genome known or predicted to be transcribed. This new generation of GeneChip allows investigation of the expression of individual exons to analyze alternative splicing. Exon Array GeneChips are available for human, mouse, and rat.Affymetrix GeneChip® System for GenotypingThe Affymetrix GeneChip system is a commercial microarray platform that allows high-throughput genotyping for human samples with as little as 250 ng of genomic DNA. The new Affymetrix® Genome-Wide Human SNP Array 6.0 contains more than 906,600 single nucleotide polymorphisms (SNPs) and more than 946,000 probes for the detection of copy number variation. The SNP Array 6.0 drives genetics forward by enabling a new study paradigm for researchers—a whole-genome approach to replication. This new approach increases the overall statistical genetic power to detect associations and eliminates other disadvantages tied to selecting a limited subset of markers in the replication phase. Affymetrix GeneChip™ CustomSeq™ Resequencing ArraysAffymetrix GeneChip CustomSeq Resequencing Arrays provides flexible custom arrays that are capable of determining up to 300 kb of unique, high-quality, double-stranded sequence. Flexible array formats of 50kb, 100kb, or 300kb are available. Sequences may cover a single contiguous region or multiple dispersed fragments, facilitating the analysis of whole genomes, multiple genes, and/or multiple organisms on a single array.
A Human Mitochondrial Resequencing array is available as a catalog product. The array interrogates the entire16 kb of the mitochondrial genome.Affymetrix GeneChip™ Whole Genome Tiling ArraysThe Affymetrix GeneChip Tiling Array Sets contain probes that cover virtually every base pair of the genome and are designed for identifying novel transcripts, mapping sites of protein/DNA interaction in chromatin immunoprecipitation (ChIP) experiments, and other experiments. Probes are tiled at 5-35 base pair resolution, depending on the organism, as measured from the central position of adjacent 25-mer oligos. Array sets are available for a variety of organisms in several different configurations. The probes for interrogating each chromosome are contained on a single array from the whole genome array set and each array is available separately.Affymetrix GeneChip™ Legacy ArraysPrevious generations of all Affymetrix GeneChips are available for users seeking to add new data to existing gene-expression datasets. Please contact us for pricing.Please contact the Microarray Resource for more information and to discuss all projects involving Affymetrix GeneChip exon, genotyping, resequencing, or tiling arrays.AffymetrixGeneChips™All Affymetrix GeneChips are available to us at special Boston Academic Consortium prices. We provide both arrays and reagents to academic users at cost. Below we have listed the prices of some of the most commonly used arrays. If you don’t see the array you are interested in, you can find out about the full catalog of arrays that are available from Affymetrix at their website (http://www.affymetrix.com/products/index.affx) and then talk with us to discuss pricing. We can also discuss custom-array strategies if no commercial array is available for your experiment.
| Array Name | Description | Array | Reagents | Labor | Total | |
| Exon 1.0 ST Array | Human , mouse, rat > 1,000,000 probesets | $400 | $200 | $350 | $950 | |
| Gene 1.0 ST Array | Human , mouse, rat Transcriptome, annotated sequences, > 27,300 probesets | $175 | $50* | $350 | $575 | |
| U133 2.0 Plus | Human Transcriptome, complete, ~ 51,000 probesets [more info] | $400 | $200 | $300 | $900 | |
| U133A 2.0 | Human Transcriptome, annotated sequences, ~ 22,500 probesets [more info] | $250 | $200 | $300 | $750 | |
| MOE430 2.0 | Mouse Transcriptome, complete, ~ 45,000 probesets [more info] | $400 | $200 | $300 | $900 | |
| MOE430A 2.0 | Mouse Transcriptome, annotated sequences, ~ 22,500 probesets [more info] | $250 | $200 | $300 | $750 | |
| ROE230A | Rat Transcriptome, annotated sequences, ~ 16,000 probesets [more info] | $325 | $200 | $300 | $825 | |
| Canine Transcriptome, ~21,600 transcripts [more info] | $275 | $200 | $300 | $775 | ||
| Xenopus laevis Transcriptome, ~14,400 transcripts [more info] | $275 | $200 | $300 | $775 | ||
| Zebrafish (Danio rerio) Transcriptome, ~14,900 transcripts [more info] | $275 | $200 | $300 | $775 | ||
| S98 | Yeast Transcriptome, ~7,000 probesets [more info] | $275 | $200 | $300 | $775 | |
| ATH1 | Arabidopsis Transcriptome, ~16,000 probesets [more info] | $275 | $200 | $300 | $775 | |
| Drosophila Transcriptome, ~13,500 probesets [more info] | $275 | $200 | $300 | $775 | ||
| C. elegans Transcriptome, ~22,500 probesets [more info] | $275 | $200 | $300 | $775 | ||
| E. coli Transcriptome, ~5,500 probesets [more info] | $275 | $100 | $300 | $675 | ||
| P. aeruginosa Transcriptome, ~6,000 probesets [more info] | $275 | $200 | $300 | $775 | ||
Reagents and LaborThe reagent fee covers our direct cost for the enzymes and materials required to prepare labeled cRNA and hybridize it to the GeneChip microarray. From start to finish, the sample labeling and hybridization takes about three days, and the labor fee covers our personnel costs for this processing. Our charge for labor is competitive with other facilities and includes assistance with data analysis, a service unique to the Boston University Microarray Resource.Experimental Design and Data AnalysisIn order to make sure that you get the most out of your microarray experiment, we provide users of the Microarray Resource with free data analysis assistance. This includes assistance with experimental design and statistical analysis, as well as access to software that allows sophisticated data mining and visualization.how to get startedAffymetrix GeneChips
- Set up a meeting with Drs. Alekseyev and Lenburg to plan your experiment, discuss RNA isolation methods, costs and/or have them order the GeneChips that you need. If you are familiar with microarray analysis, you may want to simply have us order GeneChips for you.
- Isolate either total RNA (most common) or polyA-selected RNA and deliver the sample to Dr. Alekseyev. See our list of RNA Isolation protocols.
- Wait about 3 days while the microarray resource team labels your RNA, hybridizes it to the GeneChips, collects the data using a laser scanner and presents you with copies of the results.
- Go over basic data analysis with a member of the microarray resource or on your own once you learn how to do it.
RNA Isolation ProtocolsIntroductionMany different RNA isolation protocols are compatible with the Affymetrix GeneChip and custom array sample preparation protocol. The only requirement is that you are able to isolate 5-10 micrograms of high-quality intact RNA that does not contain significant amounts of contaminating DNA.
If you currently have a preferred method for isolating RNA, continue to use it.
If you haven’t isolated RNA from your system before and your organism is not on the following list, we can help you identify established protocols that have been used for isolating RNA for Northern blotting or RT-PCR from your system. These types of protocols will also yield “microarray-grade” RNA.
Always run an agarose gel of your RNA to assess the quality.
NOTE: If you use a non column based purification reagent such as TRIzol we recommend you precipitate your RNA a second time from ethanol or you pass it through an RNeasy column or similar device. This insures removal of all organic contaminants (See spectra below).
Affymetrix recommends: If going directly from TRIzol-isolated total RNA to cDNA synthesis, it may be beneficial to perform a second cleanup on the total RNA before starting. After the ethanol precipitation step in the TRIzol extraction procedure, perform a cleanup using the QIAGEN RNeasy Mini Kit. Much better yields of labeled cRNA are obtained from the in vitro transcriptionlabeling reaction when this second cleanup is performed.RNA PurityA260 readings and 260/280 ratios are not enough to ensure high purity RNA. You must take a full UV spectrum. Below are 3 example spectra which all have good 260/280 ratios, but have very different purities. You can also use the 260/230 ratio to help determine the amount of organic contamination in your RNA. It is a much more variable number than the 260/280 ratio, but generally, ratios above 1 indicate good purity.
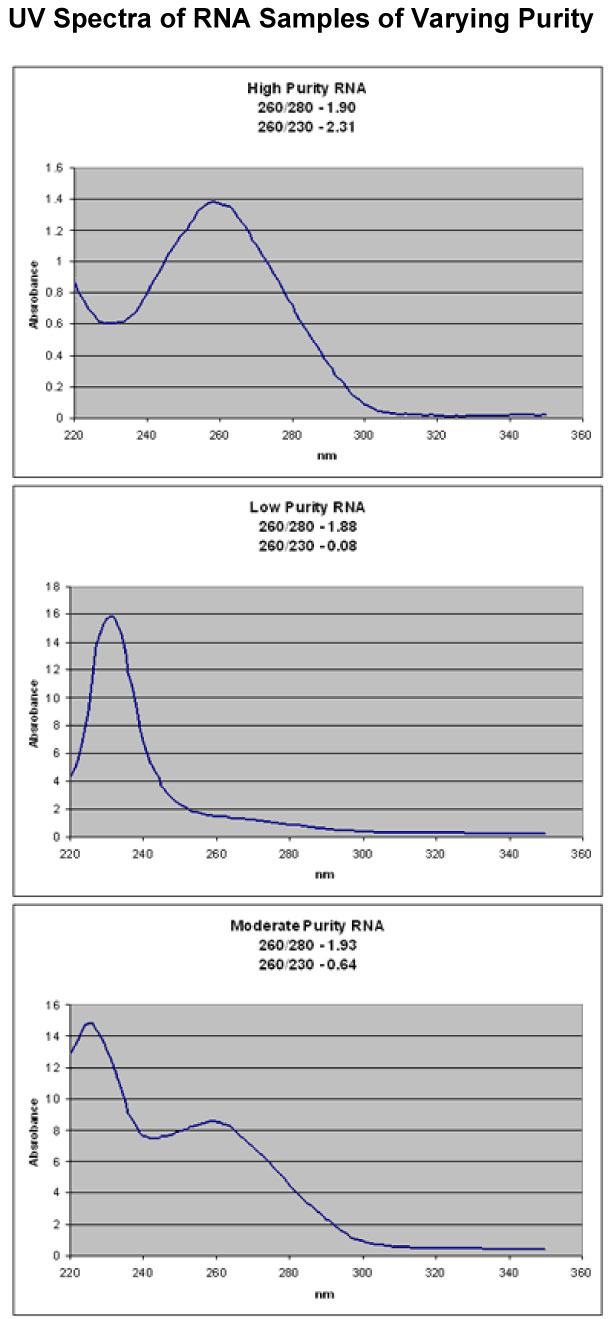
All three samples appeared clean when just taking the A260 and 260/280 ratios. It only becomes clear that they have very different purities when a full UV spectrum is taken.Yeast Total RNAGood quality total RNA from yeast cells can be obtained using the hot phenol protocol described by Schmitt, et al. (1990) Nucl Acids Res, 18:3091-3092.Hot phenol purification of RNA from yeast
- Harvest 10 ml of culture by centrifugation and resuspend in 400 ul of AE buffer (AE buffer : 50 mM Na-acetate, pH 5.3, 10 mM EDTA). Transfer to 1.5ml microfuge tube (use a tube with a screw-on cap).
- Add 40 ul of 10% SDS, vortex. Add equal volume (440 ul) phenol (Schmitt et al. say phenol should be pre-equilibrated with AE buffer but it works fine with regular Tris pH 8-equilibrated phenol).
- Vortex, incubate at 65°C for 4 min. Chill rapidly in a dry ice / ethanol bath until phenol crystals appear, centrifuge for 2 min at maximum speed.
- Transfer upper aqueous phase to fresh microfuge tube, extract with phenol / chloroform at RT for 5 min. To extracted aqueous phase add 3 M Na-acetate, pH 5.3, to final conc. of 0.3 M (i.e., add 40 ul) and 2.5 volumes of ethanol to precipitate the RNA. Wash the pellet with 80% ethanol, dry and resuspend in 20 ul of sterile water. Store at -70°.
Mammalian Cell CultureAffymetrix recommends using QIAGEN?s RNeasy Total RNA Isolation kit. Link to RNeasy product info (link opens in a new window).Mammalian TissueAffymetrix recommends using TRIzol Reagent for the initial purification of RNA from mammalian tissue. This is followed by a subsequent sample “clean-up” using QIAGEN?s RNeasy Total RNA Isolation kit (see above).
Link to TRIzol product info (link opens in a new window).
Example TRIzol purification of RNA from mammalian tissue protocolThis protocol was developed by Chuck Perou for isolating RNA from tumor specimens (C.M. Perou et al. 1999. PNAS 96: 9212-9217). This protocol may need to be modified for isolating RNA from other specimens.
- The frozen tissue specimen is removed from the freezer and, using a scalpel, small pieces (approximately 50 – 100 milligrams each) are cut off of the larger tissue specimen while it is still frozen. As the small pieces are cut off, they are immediately placed into 10 – 12mls of TRIzol Reagent (Invitrogen) that is contained within a 50ml Screw Cap Tube at RT. For a given tissue sample, upto 1 gram can be placed into this volume of TRIzol, and typically 0.5 – 1 gram of tissue is used per isolation if possible. Note, that as little as 0.2 gram has been successfully used in this volume but it may be better to use 5 – 8mls TRIzol for specimens of this size (the TRIzol Reagent Protocol calls for 1ml TRIzol per 50 – 100mg tissue processed).
- The tissue sample in TRIzol is homogenized using a PowerGen 125 Tissue Homogenizer (Fisher Scientific) starting at 5000 RPM and gradually going up to approximately 20,000 RPM over a period of 30 – 60 seconds at RT. The homogenization is performed until a homogeneous solution is obtained and very few visible tissue pieces can be seen.
- Incubate at RT for 5-10 minutes after homogenization.
- The TRIzol/tissue homogenate is transferred to a 50ml Oak Ridge Centrifuge tube and centrifuged at 12,000g (10,000RPM on SS34) for 5 – 10 minutes at 4°C.
- Remove the upper fat layer by using a Pasteur pipette hooked up to a vacuum flask (the upper fat layer, if present, has a yellowish appearance while the TRIzol homogenate remains red).
- NOTE: From here to Step 12, the protocol is exactly as written in the TRIzol Reagent Protocol.
- Add 0.2ml Chloroform per 1ml TRIzol Reagent used in Step 2. Shake tube vigorously for 15-30 seconds by hand and incubate at RT for 5 minutes.
- Spin at 12,000g for 15 minutes at 4°C. Carefully remove the upper aqueous phase, which contains the total RNA, and place this in a new 50ml Oak Ridge Centrifuge tube.
- Precipitate the RNA by adding 0.5ml Isopropyl alcohol per 1ml TRIzol Reagent used in Step 2. Incubate at RT for 10 minutes, then spin at 12,000g for 10 minutes at 4°C.
- Carefully decant the supernatant and wash the pellet once using 75% EtOH by adding 1ml 75% EtOH per 1ml TRIzol used in Step 2. Spin at 7,500g (7500 RPM in SS34) for 5 minutes at 4°C.
- Carefully decant the supernatant and air dry the pellet for 10 – 20 minutes at RT.
- Resuspend the RNA pellet in 200-300 microliter DEPC-DH20 or DEPC-TE, and take A260 if necessary.