Advancement Made in the Visualization of Large, Complex Datasets
An improvement to the premier data visualization tool t-distributed Stochastic Neighborhood Embedding (t-SNE), called optimized-t-SNE (opt-SNE), shines new light on researchers’ ability to view exactly what is in their datasets.
opt-SNE is an advancement of the widely used t-SNE created nearly 10 years ago. While t-SNE can accurately analyze approximately half a million cells in any given sample, in recent years, single cell datasets have become much larger. With opt-SNE, researchers can now visualize data from samples containing tens of millions of cells with unprecedented resolution.
The development of opt-SNE was led by Anna Belkina, MD, PhD, assistant professor of pathology and laboratory medicine.
In addition to its capacity to properly process big datasets, opt-SNE was also able to successfully visualize very small, distinct populations of cells in the blood samples tested (with each cell in these groups as rare as one in a hundred thousand of the total number of cells in the sample). Prior to opt-SNE, this accurate, large-scale visualization with simultaneous magnification of miniscule populations was not possible. “t-SNE was originally a “one-size-fits-all” algorithm, but opt-SNE computations are tailored to each individual dataset and this allows both a birds-eye and up-close view of what is in your sample. With opt-SNE, both the haystack and the needles within it can be seen,” explained Dr. Belkina, the corresponding author of the study. “It is a particularly valuable tool for the investigation of cytometry and single cell transcriptomics data”.
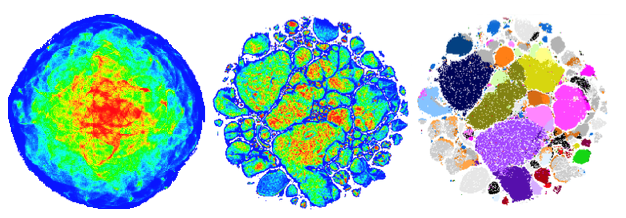 The visualization of different populations within a sample of 20 million human blood cells using t-SNE (left) and opt-SNE (middle, right)
The visualization of different populations within a sample of 20 million human blood cells using t-SNE (left) and opt-SNE (middle, right)
opt-SNE allows researchers to pinpoint previously undetectable features that distinguish diseased samples from controls. This new lens into disease states may reveal novel targets for therapies as well as new biological phenomena. This approach is already in use by multiple research groups due to Dr. Belkina’s ongoing collaborations with developers of major single cell data analysis platforms who enabled opt-SNE implementation into the Omiq.ai cloud analysis platform (Christopher Ciccolella, MS) and FlowJo software (Josef Spidlen, PhD and Richard Halpert, PhD) and co-authored the manuscript. An open-source opt-SNE package has also been released.
Additional co-authors of the study, which appears online in Nature Communications, include Rina Anno, PhD and Jennifer Snyder-Cappione, PhD.
Funding for this study was provided by the National Institutes of Health (Grant RO1AG060890-0).
Editors Note: C.O.C. is a founder of Omiq, Inc. R.H. and J.S. are employees of Beckton Dickinson (BD); FlowJo is a subsidiary of BD. The remaining authors declare no competing interests.
View all posts