Blood Microbiome
INITIATION DATE:
12.01.09
ARC DIRECTORS AND CO-DIRECTORS:
Lisa Ganley-Leal, MS, Ph.D.; Co-Director, Assistant Professor; Section of
Infectious Diseases, DOM, Dept of Microbiology
Marie McDonnell, MD; Co-Director, Assistant Professor; Section of
Endocrinology, Diabetes & Nutrition, DOM
OVERVIEW OF GOALS AND MISSION:
Abstract
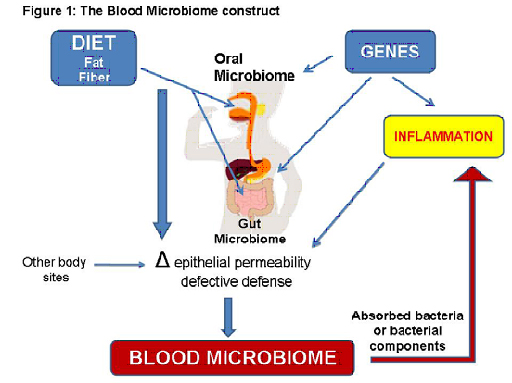
The Blood Microbiome collaborative was founded on the concept that the importance of cataloguing large human microbiomes (e.g. gut and mouth), such as the NIH Human Microbiome Project, lies within the potential for tissue microbiomes to have systemic effects (Figure 1). Our collective expertise presents a unique opportunity to advance research in this area to yield results that will likely change disease paradigms. Our effort overcomes the practical limitation that relevant diseases (e.g. diabetes and inflammatory bowel disease) do not clinically overlap to invite integrated research, despite the fact that over 200 million people worldwide suffer devastating consequences of chronic inflammatory diseases. We assert that only a committed multidisciplinary group of individuals will be able to test our hypothesis that a distinct blood microbiome exists in each, or specific clusters of chronic inflammatory diseases, and that these signature microbiomes impact the development and/or severity of disease by providing immunomodulatory ligands. All members are integral to each specific aim:
Aim 1: Detect, quantify, and qualify blood-borne bacteria in specific human cohorts to identify candidate
disease-associated microbes.
Aim 2: Define immuno-epidemiological relationships with candidate disease-associated microbes
Aim 3: Employ in vitro and in vivo models to test clinically-relevant microbes for future clinical studies.
ARC Members:
| Name/Title | Dept/School | Role in ARC |
| Lisa Ganley-Leal, M.S., PhD; Assist Prof | Section of Infectious Diseases, DOM, Department of Microbiology | Co-Director API |
| Marie E. McDonnell, MD; Assist Prof | Section of Endocrinology, Diabetes and Nutrition, DOM | Co-Director API |
| Caroline Apovian, MD, FACP, FACN; Assoc Prof | Medicine/ Pediatrics | API |
| Caroline A. Genco, PhD.; Prof | Department of Microbiology | API |
| Frank C. Gibson III, PhD; Assoc Prof | Section of Infectious Diseases, DOM | API |
| Hatice Hasturk, DDS, PhD; Adjunct Assoc Prof | Department of Periodontology and Oral Biology/Goldman School of Dental Medicine | API |
| Alpdogan Kantarci, DDS, PhD; Assoc Prof | Department of Periodontologyand Oral Biology/ Goldman School of Dental Medicine | API |
| David J. Ecker, PhD | IBIS technologies, Abbot | API |
| Bruce J. Paster, PhD. | Forsyth Institute | API |
MAIN ARC PROJECT(S) FOR 2009-2010:
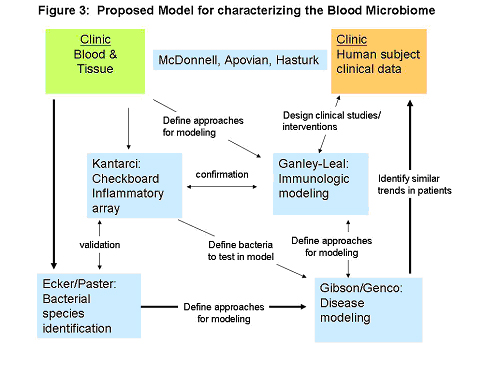
Periodontal disease, as a model of compromised oral epithelium, will be assessed as a likely promoter of chronic bacterial translocation. Blood will be drawn before and after controlled gingival manipulation (teeth cleaning, established as a cause of transient detectable bacteremia in patient cohorts. Blood samples will be distributed to the team for analyses. Clinical data will be collected and assessed in relation to inflammatory signatures and identified bacterial species. Blood/tissue samples and information flow between each discipline is represented in Figure 3. Through this pathway we will complete a series of proof-of-concept studies and model development to test the hypothesis that the presence of microbes in blood initiates and/or propagates the chronic inflammatory state that accompanies many noninfectious inflammatory diseases, with an initial focus on the highly prevalent disease of obesity-related T2DM.
ARC AS A RESOURCE:
Our group has been collaborating to define how chronic inflammatory disease changes system immune responses with little dedicated funding. In addition to providing seed funding, the ARC will allow us to generate sufficient preliminary data to apply for a Program Project Grant to characterize the Blood Microbiome in many chronic inflammatory diseases including inflammatory bowel disease, obesity, HIV, cardiovascular and periodontal disease. Thus, we anticipate that the PPG will include several other faculty at BU who perform research on these diseases, including Frank Farraye in Gastroenterology, Noyan Gokce in Cardiology, Paul Skolnik in Infectious Disease and Tom Van Dyke in the School of Dental Medicine.
Additionally, the Blood Microbiome ARC will intersect with the proposed Inflammation Training Program (PI Genco, Co-Is Gibson, Ganley-Leal, McDonnell). This Institutional T32 training program will provide predoctoral and postdoctoral trainees with a solid academic background in immunology with emphasis on multidisciplinary approaches to study common mechanisms of inflammation.
B.2.c: Envisioned application of technologies in the ARC : Since conventional methods for detecting low levels of bacteria in the blood have several limitations16, we will employ three methods for sensitive bacterial DNA detection in blood in order to define an ideal and most practical Model for investigating the Blood Microbiome: 1) PCR/ESI-MS technology, 2) HOMIM, and 3) Checkerboard analysis (see above Bacterial Genome Detection and Identification).
B.3.d Plans for frequency of meeting and self-evaluation: Because the amount of data generated from this collaboration will be significant, we will have monthly, scheduled ARC meetings to present data and directly address rational directions of the project. We plan to have each study area (e.g. Clinical, Immuno-epidemiology) present on a rotating basis. This structure will be supported by continuing our current practice of frequent (often daily) communication between coinvestigators, and informal interactions for clarifications and idea-sharing. Non-institutional investigators will be available by conference call and will travel as needed to clarify results and refine interpretation.
B.3.e Potential Collaborations with outside institutes: We have already incorporated two collaborations from outside institutions. Further, we are in the infancy of establishing collaborations on pediatric patients with chronic inflammatory disease. For example, we are initiating studies at MGH on pediatric inflammatory bowel disease but currently lack dedicated resources. Our work may therefore inform us of the etiology of Blood Microbiome and the ultimate design of rational interventions in children for prevention of disease.
Pictures, Images, Figures: