COVID-19 Response – for basic science researchers
11
General:
In the midst of this pandemic, we in the Center for Regenerative Medicine (CReM) of Boston University and Boston Medical Center recognize both the urgency to gain a greater understanding of the pathophysiology of COVID-19, and the importance of establishing a model that can be used to study SARS-CoV-2 (the virus that causes COVID-19) and adapted to test potential therapeutic candidates. Using our knowledge of developmental biology, our research center has developed protocols to differentiate human induced pluripotent stem cells (iPSCs) into both alveolar and airway epithelial cell lineages. Importantly, we observe the expression of ACE2 and TMPRSS2, two host cell proteins that have been shown to be required for SARS-CoV-2 cell entry (Hoffmann et al., Cell, 2020), in our iPSC-derived cells, raising the possibility of using our iPSC-based systems to model SARS-CoV-2 infection in vitro. If you are interested in collaborating and utilizing our system, please refer to the details of our alveolar and airway model below.
Instructions regarding requests for our cells, collaborations, or assistance can be found at the end of the page. If you choose to refer to our unpublished data in urgent grant applications for COVID research, please let us know and credit: Laboratories of Darrell Kotton, Andrew Wilson, and Finn Hawkins, Center for Regenerative Medicine (CReM) of Boston University and Boston Medical Center, Boston, MA, USA. Letters of support are available upon request. For non-urgent applications or those unrelated to COVID, please first contact us for permission. Additional detailed protocols are posted on the protocols page of our CReM website.
**For the latest analysis of our iPSC-based model, please feel free to check our recently posted BioRxiv Preprint!
iPSC-derived alveolar epithelium model:
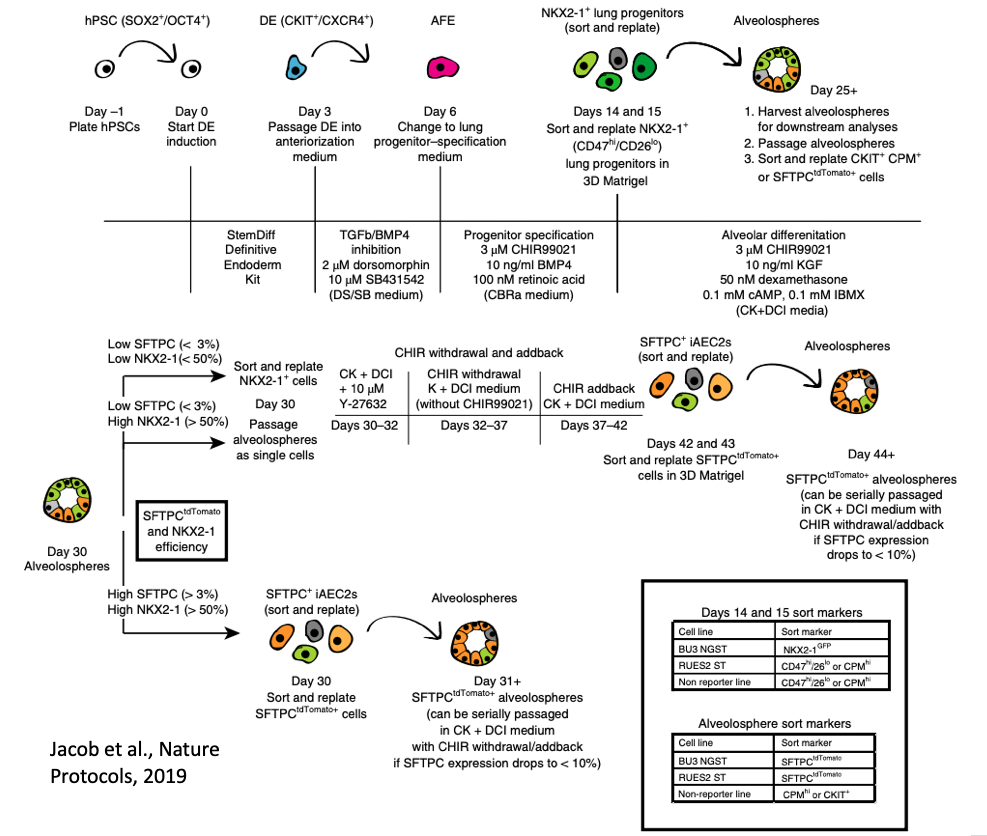
Primary human lung alveolar epithelial cells are challenging to obtain and maintain in culture. By manipulating the growth factors in media and utilizing reporters for NKX2-1 (marking lung fate) and SFTPC (marking distal lung and alveolar epithelial type II cells, AEC2s), we have developed a differentiation protocol (shown above and accessible here) to generate human iPSC-derived AEC2s (iAEC2) that can be maintained and passaged in 3D culture (Jacob et al., Cell Stem Cell, 2017; Jacob et al. Nature Protocols, 2019).
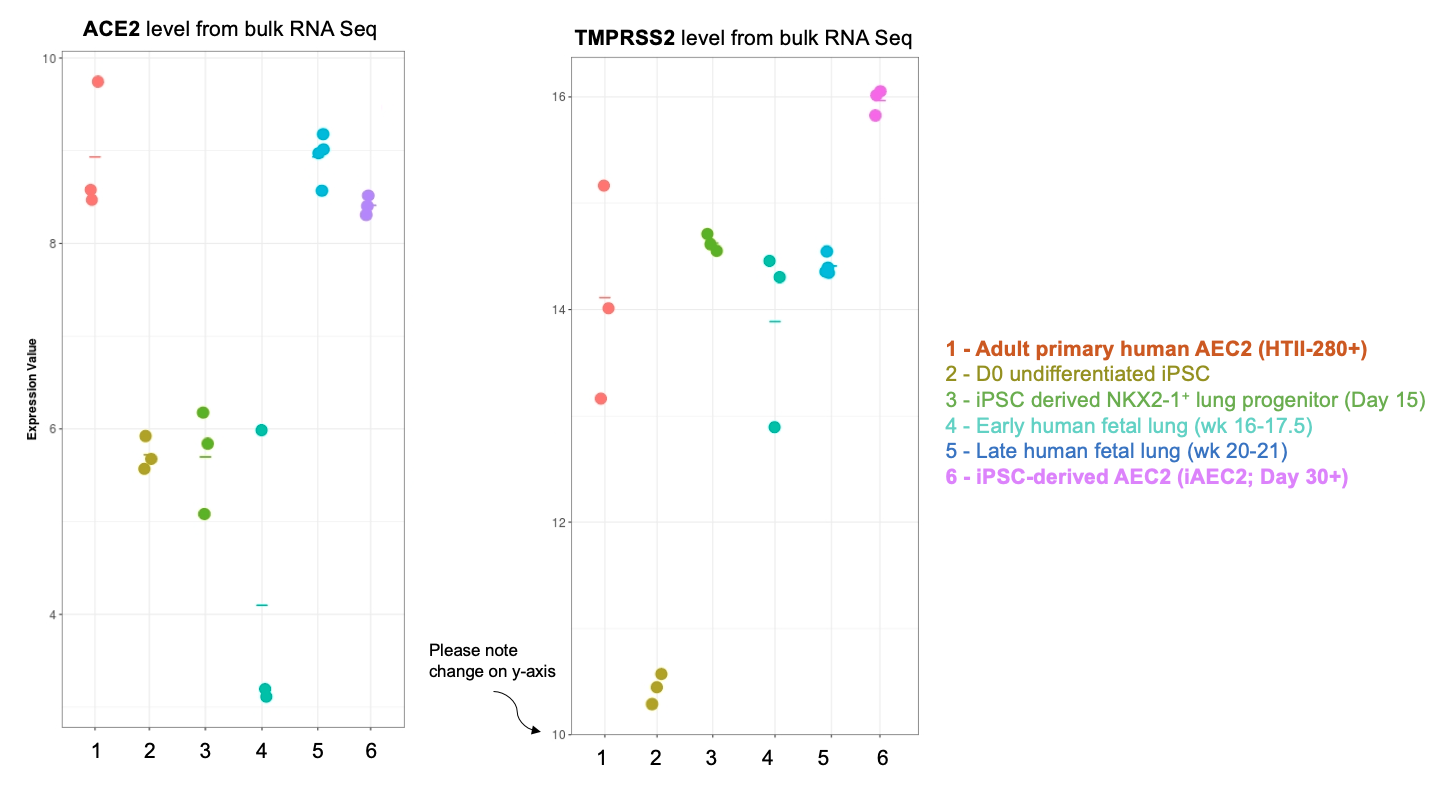
Importantly, our bulk RNA Sequencing data has shown that our iAEC2 spheres have similar expression levels of ACE2 and TMPRSS2, two essential host proteins for SARS-CoV-2 cell entry (Hoffman et al, Cell 2020), compared to primary adult human freshly purified AEC2s, sorted from three different adult lungs using the HTII280 antibody (unpublished data below as dot plot figure).
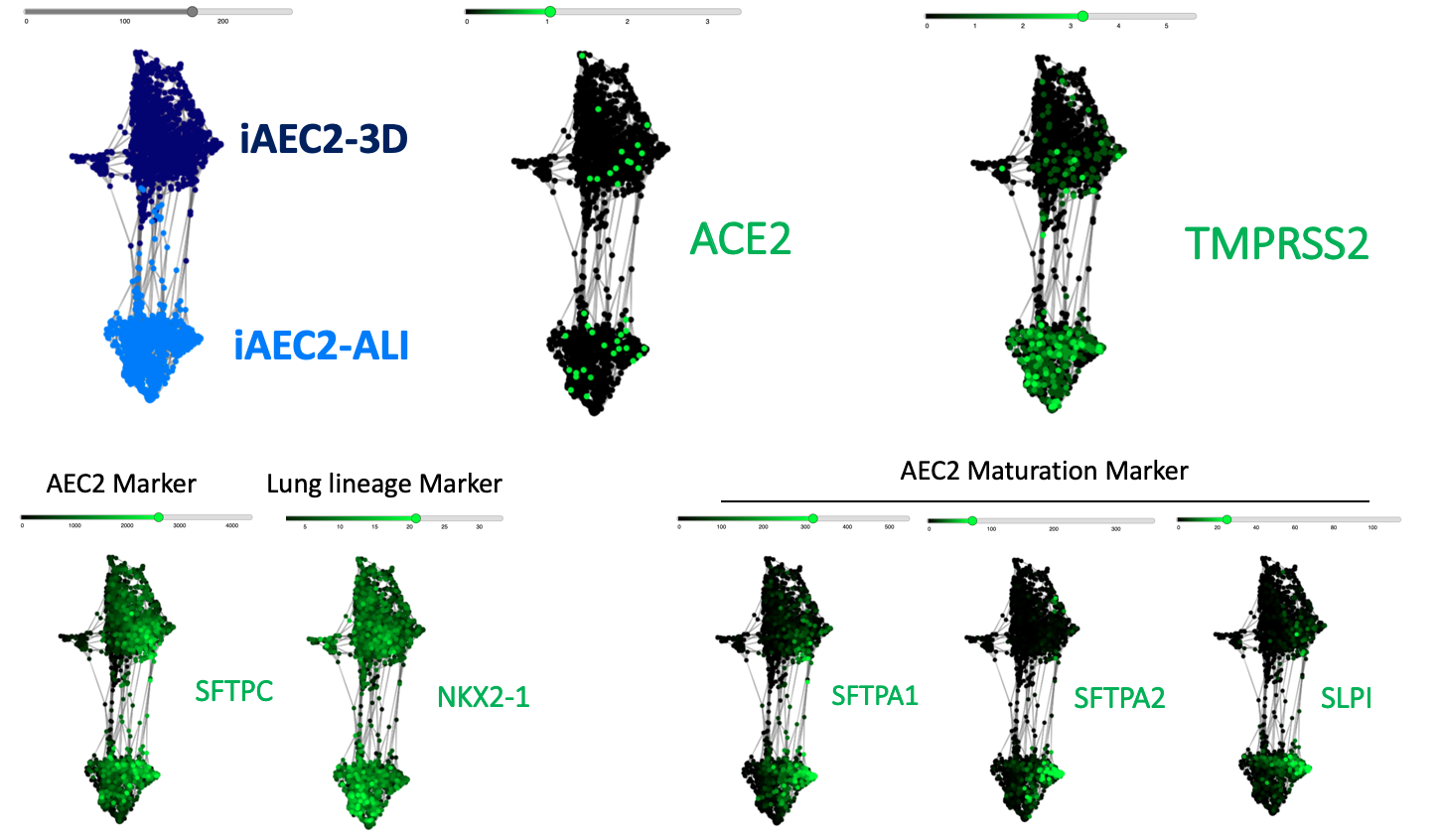
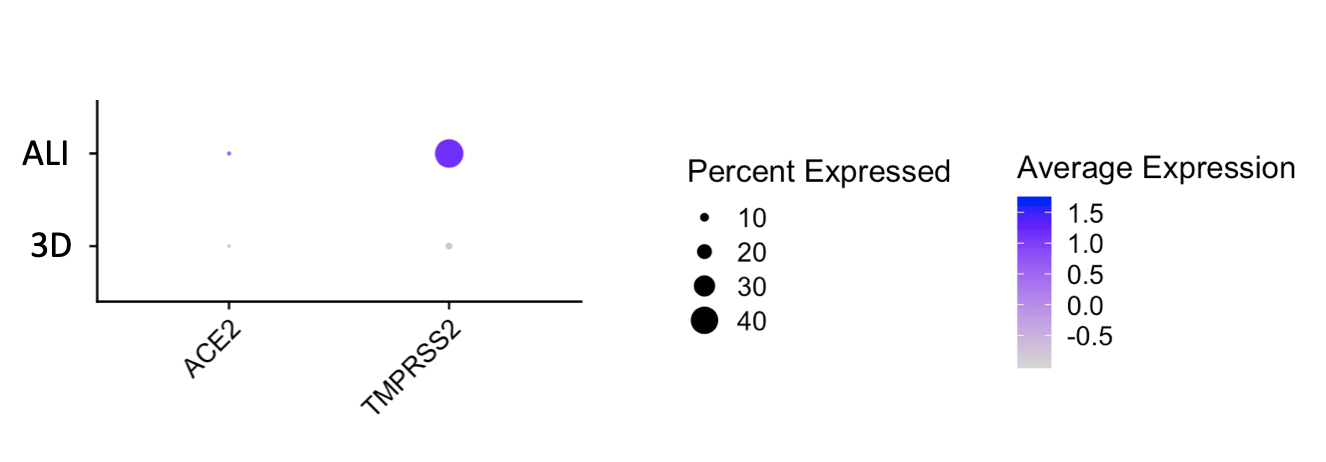
The CReM’s Wilson lab has further expanded the protocol to culture iAEC2s at air-liquid interface (ALI, unpublished data and protocol summary below). ALI culture of iAEC2s permits the exposure of SARS-CoV-2 to the apical surface of the cells. Most importantly, scRNA-seq analysis of iAEC2-ALI has revealed expression of ACE2 and TMPRSS2 (unpublished data below).
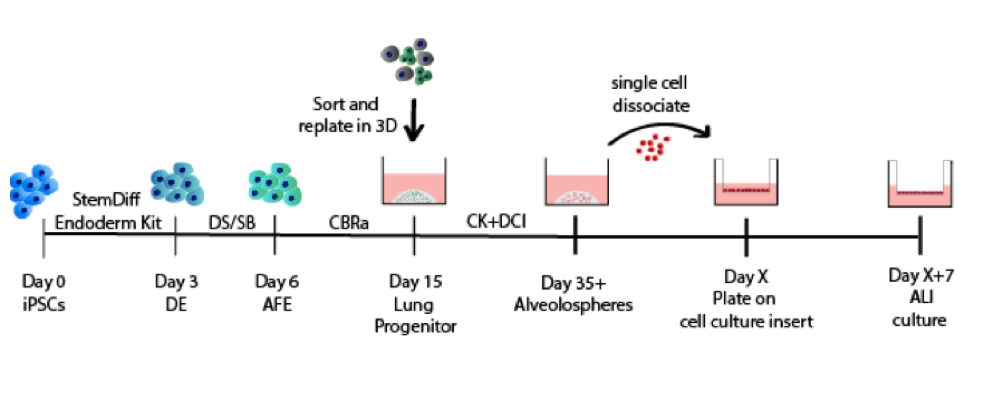
Although our differentiation protocol takes 40+ days, we will share differentiated iAEC2s as 3D self-renewing spheres in live cell culture to save you time. Please refer to contact information at the end of the page for requests for live cultures of our cells.
iPSC-derived airway epithelium model:
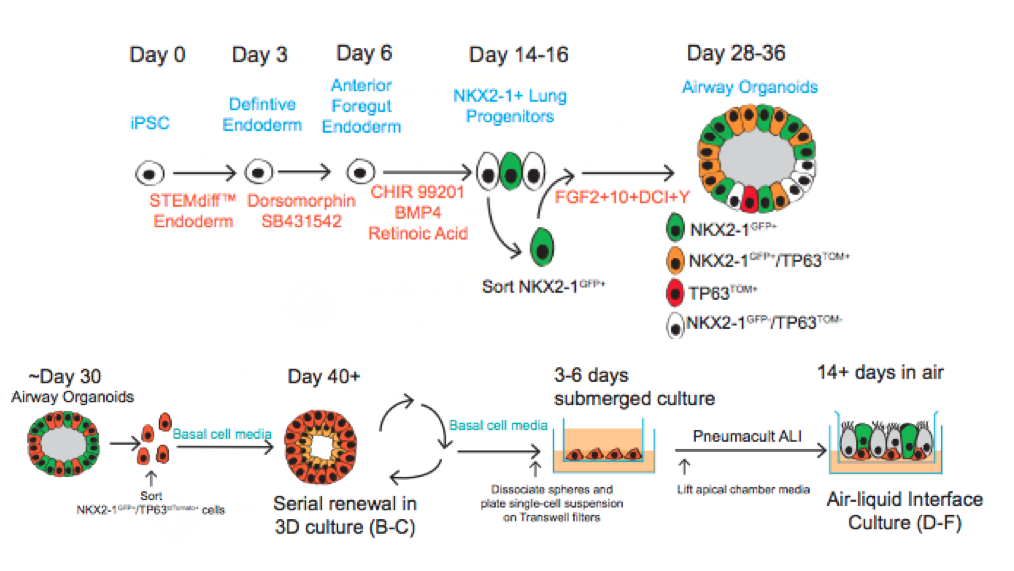
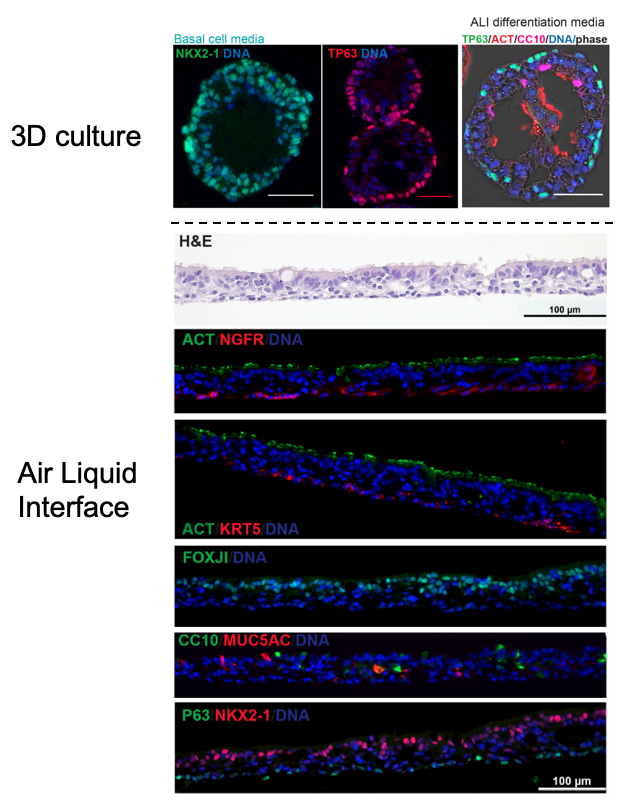 Similar to our iAEC2 system, the CReM’s Hawkins lab, in collaboration with the Brian Davis Lab in University of Texas, developed a differentiation protocol (shown above) to make human iPSC-derived airway basal cells (iBC), and maintain and passage them in 3D culture (Hawkins et al., BioRxiv, 2020). iBCs grown in differentiation media in 3D culture or plated onto 2D ALI give rise to airway epithelial lineages, including secretory cells (marked by CC10 (SCGB1A1)/ MUC5AC), multiciliated cells (marked by FOXJ1 and Acetylated Tubulin; ACT) and basal cells (marked by KRT5/ TP63/NGFR). The iPSC-derived airway epithelium can recapitulate in vivo functionality such as CFTR-mediated chloride transport, or mucus cell metaplasia with interleukin-13 exposure (Hawkins et al., BioRxiv, 2020).
Similar to our iAEC2 system, the CReM’s Hawkins lab, in collaboration with the Brian Davis Lab in University of Texas, developed a differentiation protocol (shown above) to make human iPSC-derived airway basal cells (iBC), and maintain and passage them in 3D culture (Hawkins et al., BioRxiv, 2020). iBCs grown in differentiation media in 3D culture or plated onto 2D ALI give rise to airway epithelial lineages, including secretory cells (marked by CC10 (SCGB1A1)/ MUC5AC), multiciliated cells (marked by FOXJ1 and Acetylated Tubulin; ACT) and basal cells (marked by KRT5/ TP63/NGFR). The iPSC-derived airway epithelium can recapitulate in vivo functionality such as CFTR-mediated chloride transport, or mucus cell metaplasia with interleukin-13 exposure (Hawkins et al., BioRxiv, 2020). 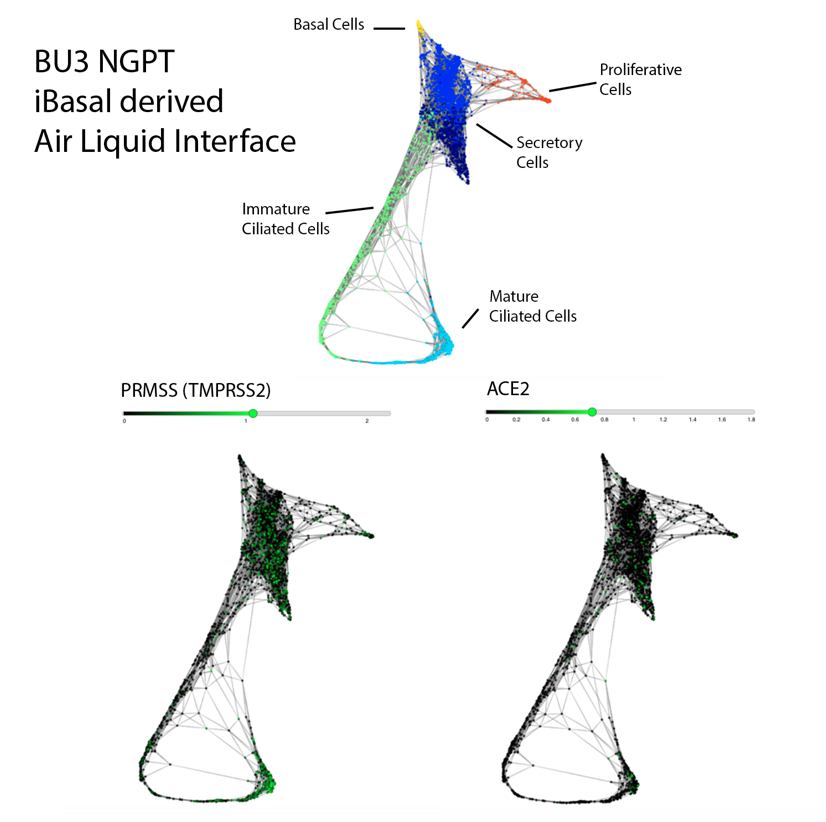
Importantly, scRNA-seq analysis of iBC-derived airway ALI cultures after differentiation (unpublished data shown above) demonstrates expression of ACE2 and TMPRSS2, in the secretory, ciliated and basal cell lineages, raising the possibility of modeling the effect of SARS-CoV-2 on airway epithelium (View and interact with unpublished scRNA-seq dataset via SPRING here).
Although the differentiation protocol usually requires 50+ days, we can share our iBCs in 3D sphere live cultures or as frozen vials. Please refer to contact information at the end of the page.
Comparison of our iPSC-based systems to primary adult human distal lung at single-cell resolution
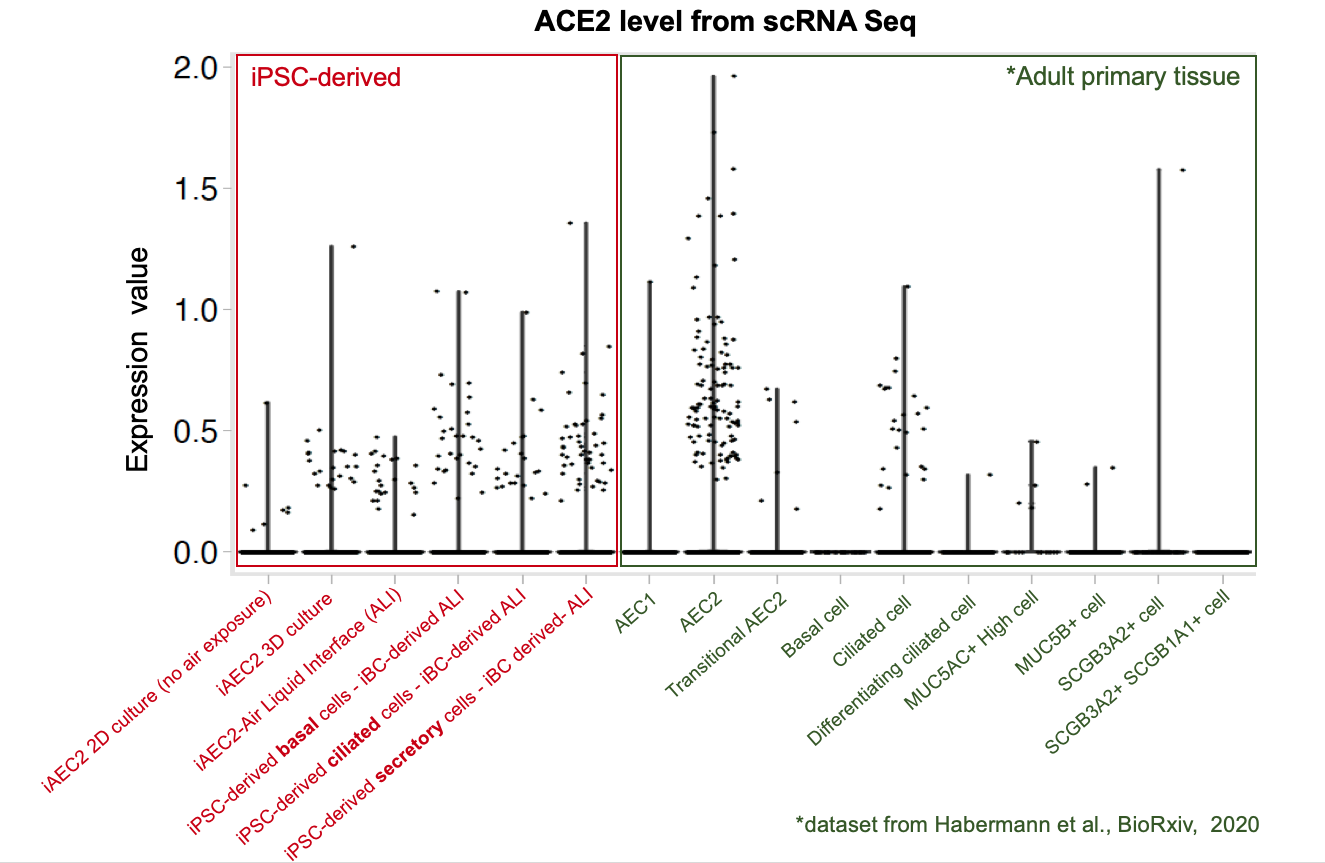
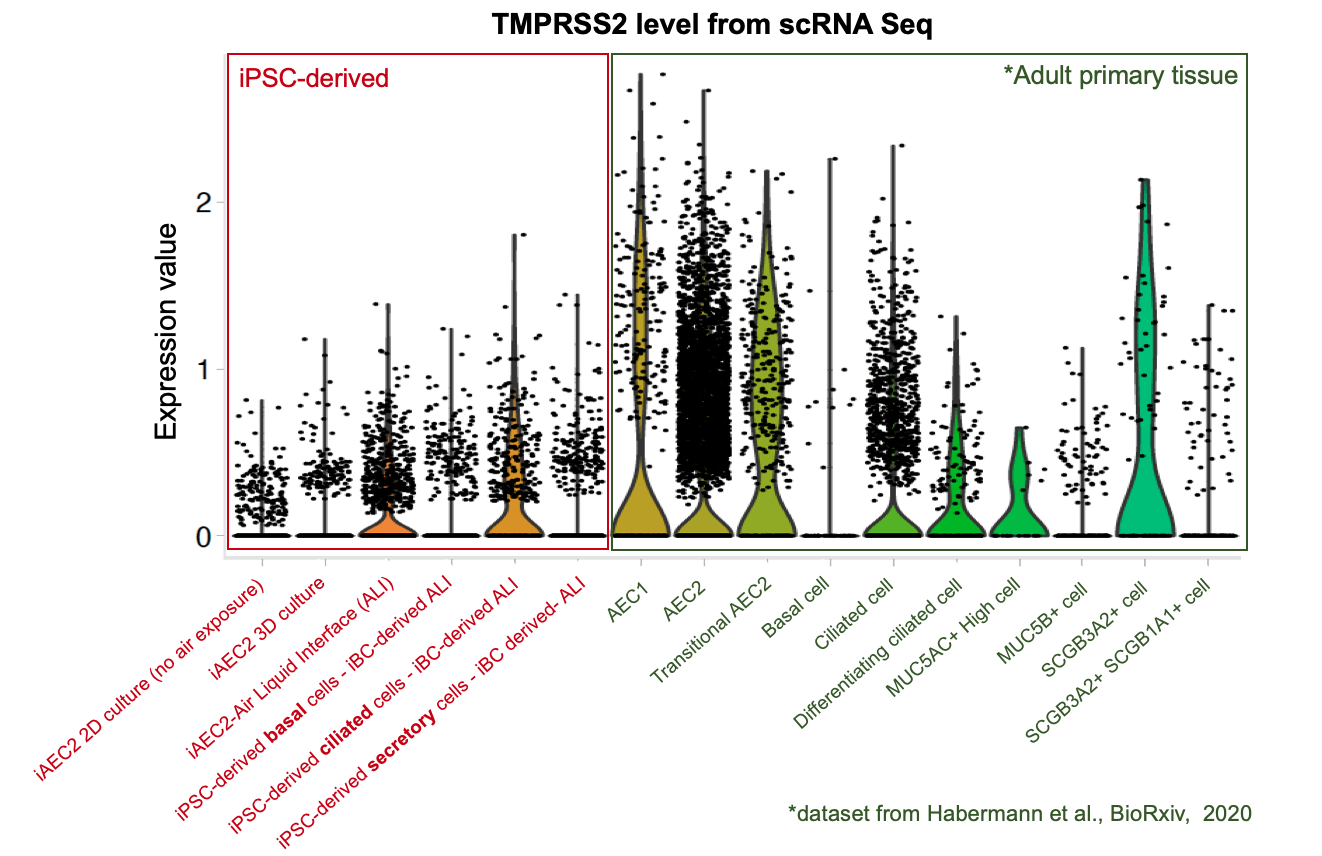
To further evaluate whether our iPSC-based systems recapitulate the expression of ACE2 and TMPRSS2 in each specific cell lineages in the airway and alveoli, we analyzed our scRNA-seq datasets together with an adult primary human lung scRNA-seq dataset (Habermann et al., BioRxiv, 2020). Quantitative analysis (shown in the violin plots above) suggests that our iPSC-derived cells express ACE2 and TMPRSS2 at levels close to their primary counterparts in the adult human lung . This further suggests the potential of our iPSC-based airway and alveolar systems as in vitro models for COVID-19.
Instructions to request our assistance:
If you would like to work with our iPSC-based models, please click here for details regarding putting in a request for our cells.
If you would like to pursue a collaboration or request a letter of support, please contact our administrative assistant Meaghan Tetro (mtetro at bu.edu)
To reference these data, please credit: Online data posted online at www.kottonlab.com courtesy of the Laboratories of Darrell Kotton, Andrew Wilson, and Finn Hawkins, Center for Regenerative Medicine (CReM) of Boston University and Boston Medical Center, Boston, MA, USA
If you would like to make a donation to financially support our cell sharing mission, click here!