S. Thiagalingam, PhD

Sam Thiagalingam, Ph.D.
Associate Professor of Medicine and Pathology & Laboratory Medicine
Ph.D. The Johns Hopkins University
M.S. Bowling Green State University
B.Sc. (Hons.) University of Jaffna
Post-doctoral Fellowship, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins
Contact:
Phone: (617)-638 6013; Fax: (617)-638 4275
E-mail: samthia@bu.edu;
Web: http://people.bu.edu/samthia/index.html
Elucidation of the molecular basis of cancer metastasis and discovery of biomarkers for diagnosis, prognosis and therapy of cancer and psychiatric disorders
Our major research focus is on the use cancer genomics, employing primarily breast and colon cancers as model systems, to shed light on genomic instability, genetic and epigenetic aberrations and metastasis of cancer. We hope to elucidate the molecular basis of the multi-step cancer progression through these studies. Furthermore, we are also interested in unravelling the correlation between the epigenome and the pathogenesis of major psychiatric disorders such as schizophrenia (SCZ) and bipolar disorder (BD).
Our pioneering studies analysing the LOH frequencies of colon cancer showed that SMAD4 is the major target tumor suppressor gene localized to the minimally lost region on chromosome 18q. Subsequent studies have validated these observations in tumor analyses and from the comparative analysis of mouse models. As a follow up to these observations, based on our preliminary studies, we plan to test the hypothesis that the direct/indirect inactivation of Smad4 is a major switch allowing the conversion of benign tumors to the metastatic form during colon cancer progression.Except for the elucidation of an association between genetic alterations in the SMAD4 gene and gastrointestinal and pancreatic cancers, the nature and contributions of the other SMAD gene alterations in cancers is largely unknown. Therefore, we developed a novel technique known as Targeted Expressed Gene Display (TEGD) to survey various SMAD genes for differential expression. The loss of SMAD8 expression in multiple types of cancers, including 31% of both breast and colon cancers directly correlated with epigenetic silencing by DNA hypermethylation. We are in the process of investigating the temporal relationship between epigenetic inactivation of the SMAD8 gene and the stage(s) of breast cancer and are planning to determine the identities and roles of differentially regulated genes due to defective Smad8 signaling that mediate genesis and metatstasis of breast cancer.
We have also continued to maintain an interest in understanding the connection between genomic instability and cancer at the molecular level. It has been universally believed that defective spindle assembly checkpoint (SAC) causes aneuploidy. Interestingly, our studies uncovered a novel apoptotic checkpoint pathway regulated by the kinetochore proteins. We plan to continue these studies by performing a detailed analysis of the roles of individual kinetochore proteins in the maintenance of genomic stability.
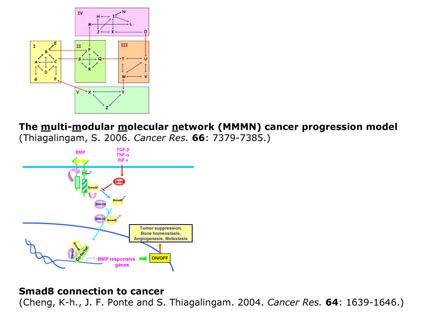
Our studies involving genetic and epigenetic analysis of lung cancer and the examination of the literature in the cancer field have led us to propose an academically simplified scheme to explain the complexity in cancer progression as a process that consists of a series of a cascade of interconnected functional sub-network modules of various alterations in a multi-modular molecular network (MMMN) encompassing multiple targets within each module. Our long-term goal is to contribute to the elucidation of the multi-modular molecular network (MMMN) cancer progression models as the road map to dissect the complexity inherent to cancer. Currently, we are in the process of using breast cancer as a prototype to perform proof of principle analyses to construct a MMMN cancer progression model.
The fact that genetic makeup alone cannot explain the molecular basis of major psychiatric disorders such as SCZ and BD was instrumental for us to direct our attention to epigenetic alterations as the major cause of pathogenesis. Our studies showed that both hyper- as well as hypo- promoter DNA methylation changes of the genes RELN and MB-COMTrespectively play critical roles in defining their altered functionality in major psychiatric disorders, SCZ and BD. Therefore, because of the possibility of gene-environment interactions mediated epigenetic modulation of gene function, we are involved in establishing a logical relationship between epigenetic changes and schizophrenia and bipolar disorder.
Selected Publications:
Thiagalingam, S., and D.V. Faller 2008. The Cancer Epigenome: Can it be targeted for therapy? In Molecular Targeting in Oncology, H. L. Kaufman, S. Wadler and K. Antman (Eds). Humana Press Inc., Chapter 5: 97-113.
Thiagalingam, S., and P. Papageorgis. 2008. DNA methylation profiles as prognostic markers for cancer. In Cancer Epigenetics, T. Tollefsbol (Ed) CRC Press (Taylor and Francis), Chapter 19: 335-348.
Abdolmaleky, HM., C.L. Smith, J-R. Zhou and S. Thiagalingam. 2008. Epigenetic Alterations of the Dopaminergic System in Major Psychiatric Disorders. Pharmacogenomics in Drug Discovery and Development. In Methods in Molecular Biology. Q. Yan and J. Walker (Ed). Humana Press Inc., Totowa, NJ., 448: 187-212.
Pan, H., F. Gao, P. Papageorgis, H. Abdolmaleky, D.V. Faller and S. Thiagalingam. 2007. Aberrant activation of g-catenin promotes genomic instability and oncogenic effects during tumor progression. Cancer Biology & Therapy 6:1638-43.
Thiagalingam, S. 2006. A cascade of modules of a network defines cancer progression . Cancer Res. 66: 7379-7385.
Abdolmaleky, HM., K-h. Cheng, S.V. Faraone, M. Wilcox, S. J. Glatt, F. Gao, C.L. Smith, R. Shafa, B. Aeali, H. Pan, P. Papageorgis, J.F. Ponte, V. Sivaraman, M. Tsuang and S. Thiagalingam. 2006. Hypomethylation of MB- COMT Promoter is a major risk factor for Schizophrenia and Bipolar Disorder. Human Mol. Genet. 15: 3132-3145.
Russo, A. L., A. Thiagalingam, H. Pan, J. Califano, K-h. Cheng, J.F. Ponte, D. Chinnappan, P. Nemani, D. Sidransky, and S. Thiagalingam. 2005. Differential DNA hypermethylation of critical genes mediate the stage specific tobacco smoke induced neoplastic progression of lung cancer. Clin. Cancer Res. 11: 2466-2470.
Pan, H., J. Califano, J. F. Ponte, A.L. Russo, K-h. Cheng, A. Thiagalingam, P. Nemani, D. Sidransky and S. Thiagalingam. 2005. Loss of heterozygosity patterns provide fingerprints for genetic heterogeneity in multistep cancer progression of tobacco smoke-induced non-small cell lung cancer. Cancer Res. 65:1664-1669.
Abdolmaleky, H., K-h. Cheng, A. Russo, C.L. Smith, S.V. Faraone, R. Shafa, M. Wilcox, S. Glatt, W.S. Stone, G. Nguyen, J.F. Ponte, S. Thiagalingam and M. Tsuang. 2005. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: A preliminary report. Am J Med Genet B Neuropsychiatr Genet. 134B: 60-66.
Cheng, K-h., J. F. Ponte and S. Thiagalingam. 2004. Elucidation of epigenetic inactivationof SMAD8 in cancer using Targeted Expressed Gene Display. Cancer Res. 64: 1639-1646.
Thiagalingam, S., R. L. Foy, K-h.Cheng, H. J. Lee, A. Thiagalingam, and J. F. Ponte. 2002. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Current Opinion in Oncology 14: 65-72. (Correction: 14(3): 374).
Thiagalingam, S., S. Laken, J. K. V. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein and C. Lengauer. 2001. The mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. USA. 98: 2698-2702.
Thiagalingam, S., C. Lengauer, F. S. Leach, M. Schutte, S. A. Hahn, J. Overhauser, J. K. V. Willson, S. Markowitz, S. R. Hamilton, S. E. Kern, K. W. Kinzler and B. Vogelstein. 1996. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nature Genet. 13: 343-346.