Biochemistry Russek Day Winners
The Department of Biochemistry Henry I. Russek Achievement Day Awards Committee is pleased to announce this year’s award winners. Nathan Kingston [Dr. Xaralabos (Bob) Varelas’s lab] is the first prize winner, and Joseph Kern [Dr. Xaralabos (Bob) Varelas’s lab] is the second prize winner. Our congratulations go out to the winners as well as their advisor! We look forward to hearing about their work on Henry I. Russek Achievement Day; remember it’s this week (Thursday, May 6th and Friday, May 7th) so mark your calendars for the virtual events.
The keynote address will be delivered on Thursday May 6th at 9:20 AM by Dr. Daniel Haber, Director of the MGH Cancer Center, “Circulating Tumor Cells: Insights into Cancer Metastasis”. Please go to the following site for updated information and to register so as to get the Zoom link:
https://www.bumc.bu.edu/gms/academics/phd-programs/russek-student-achievement-day/
Research News: Lung precancer pathology
A recently published study from the Varelas laboratory has identified new mechanisms contributing to the development of precancer lesions in lung airways. Precancer lesions that arise in airways are associated with chronic lung damage, such as that induced by cigarette smoke, and such lesions are predisposed to transitioning into lung squamous cell carcinoma, a poorly treatable subtype of non-small cell lung cancers.
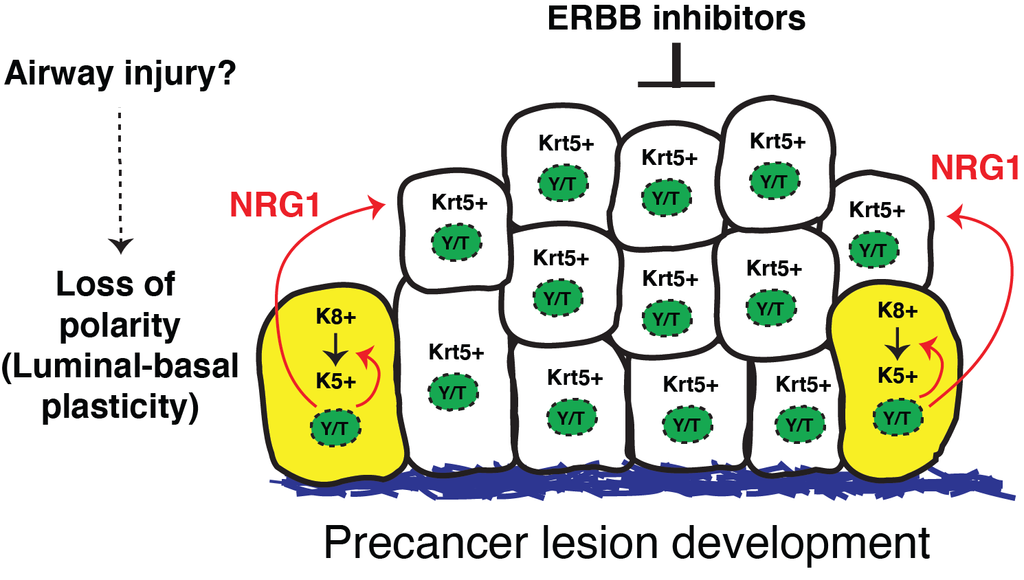
The study shows that disrupted epithelial polarity leads to airway epithelial stem cell expansion, resulting in pathologies that resemble human precancer. Aberrant epithelial polarity was induced by the conditional deletion of the polarity regulator Crumbs3, which the group showed leads to activation of the transcriptional regulators YAP and TAZ. The study showed that YAP/TAZ-regulated gene expression drives a gene expression program that stimulates epithelial plasticity and stem cell growth, and associates with the progression of precancer lesions in human patients. Through analysis of YAP/TAZ-regulated genes, an important role was uncovered for the growth factor Neuregulin-1, which is directly induced by YAP/TAZ and stimulates signaling via the ERBB-receptors. Notably, the study showed that inhibition of ERBB receptors was capably of preventing and treating precancer lesions in polarity-defective animals.
The study, which was published on April 26, 2021 in PNAS, offers new knowledge into the etiology of lung precancer pathology, and provides directions for identifying and intercepting lung squamous cell carcinoma at the earliest stages.
CBMS Receives Massachusetts Life Science Center Grant
The Center for Biomedical Mass Spectrometry (CBMS) has been awarded a grant by the Massachusetts Life Science Center to advance the ability to characterize spike protein glycosylation as respiratory viruses evolve. The CBMS applies mass spectrometry methods to meet the emerging needs in biomedicine. As part of this award, CBMS has just installed a new Waters SELECT SERIES Cyclic IMS system. The Cyclic system represents a breakthrough technology that will facilitate rapid and accurate characterization of virus spike protein glycosylation to support virus surveillance and vaccine development.
Respiratory viruses including influenza and coronaviruses evolve rapidly as they circulate in the human population. These proteins are coated with a spike protein that recognizes receptors in host airway cells. Spike proteins are decorated by sugar molecules known as glycans. These glycans enhance the structures and functions of proteins, including the virus spike proteins. Researchers track the genetic sequences of viruses as they evolve. The genetic sequences do not predict the manner in which glycans alter the structure and function of virus spike proteins.
Research News: New Discovery from the Garcia-Marcos Laboratory
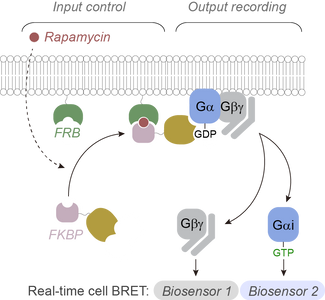
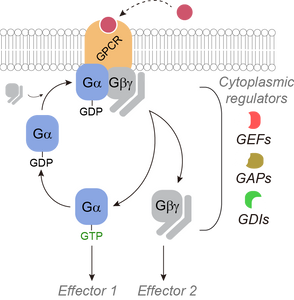
The Garcia-Marcos laboratory has discovered that heterotrimeric G-proteins, which are critical molecular switches in cellular communication processes, can trigger different responses in cells depending on the type of protein that activates them.
Heterotrimeric G-proteins are typically activated by G-protein-coupled receptors (GPCRs), which are the target for over one-third of drugs approved for use in the clinic. But G-proteins can also be activated by other proteins that are not GPCRs, a mechanism with important implications for human physiology and disease. The Garcia-Marcos laboratory has used an innovative approach to dissect and compare the consequences of G-protein stimulation by different activators, including GPCRs and various non-GPCR proteins. The approach leveraged a recently developed type of biosensors capable of detecting different forms of active G-proteins in living cells in real time. These were combined with engineered G-protein activator constructs that could be turned on at will with an exogenous synthetic chemical.
The main conclusion is that, contrary to previous beliefs, GPCR and non-GPCR activators elicit different forms of G-protein activation in cells. These findings have important implications in our understanding of pharmacologically actionable signaling hubs in cells, which could be leveraged to envision and design new therapeutic agents.
You can read the article here:
Garcia-Marcos M. Complementary biosensors reveal different G-protein signaling modes triggered by GPCRs and non-receptor activators. Elife. 2021 Mar 31;10:e65620. PMID: 33787494 https://pubmed.ncbi.nlm.nih.gov/33787494/
Grant Award: Department of Defense, Discovery Award

Robert Mercer, a Postdoctoral Associate in the Harris Lab, was recently awarded a Discovery Award research grant funded by the Department of Defense. This two-year, $200,000 grant is entitled “ Identification of Receptors for TAR DNA-binding Protein 43 (TDP-43) That Mediate Cellular Uptake and Neurotoxicity ”. This project aims to identify the cellular receptors of the pathological TDP-43 assemblies found in the brains of patients with Amyotrophic Lateral Sclerosis (ALS), Frontotemporal Lobar Degeneration (FTLD), and Chronic Traumatic Encephalopathy (CTE). The identification of these receptors will provide novel therapeutic targets to combat these devastating and currently untreatable diseases.
Dr. Grishok Receives 2021 Shipley Pilot Grant
Dr. Alla Grishok, Associate Professor of Biochemistry has been awarded a 2021 Shipley Prostate Cancer Research Pilot Grant Award. She will act as coordinator of a multi-center, multi-PI initiative involving Dr. Christopher Heaphy (Department of Medicine), Dr. Joshua Campbell (Department of Medicine), and Dr. Rachel Flynn (Department of Pharmacology and Experimental Therapeutics, GSI) across Boston Medical Center (BMC) Pathology department, Shipley Prostate Cancer Research Center, BU-BMC Cancer Center, and Genome Science Institute (GSI) at BUSM.
The goals of this pilot research are: "to (1) identify coding and non-coding genomic alterations that are associated with more aggressive disease in Black men with prostate cancer and to (2) establish a pipeline that will integrate the genomic data with other biomarker measurements, such as the presence of circulating tumor DNA or alterations in the gut microbiome, and with the overall clinical outcomes in a large cohort of men". The success of this project may lead to better predictions of the severity of the disease in individual men and to informed selection of the appropriate treatment options in the future.
Research news: Methods to improve quantitative glycoprotein coverage
Most cell surface and extracellular matrix proteins are glycosylated as a mechanism driven by evolution that elaborates their adhesive interactions with binding partners. Such glycosylation occurs by biosynthetic reactions in the secretory pathway that gives rise to heterogeneity at each glycosylation site. This heterogeneity diversifies the range of interactions and therefore functions of glycoproteins. But, how can we understand the functional roles of glycoproteins if they are heterogeneous? Conventional proteomics methods have been adapted to quantify protein glycosylation heterogeneity, but understanding glycoprotein function requires improvements in sensitivity and selectivity. Chang and Zaia describe current methods of glycosylation quantification and show the promise of emerging glycoproteomics methods that employ data-independent analysis and ion mobility separations.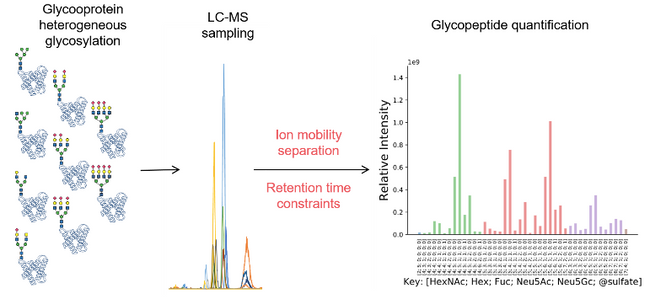
Grant Awards: Visualizing Alzheimer’s Disease
 Ladan Amin an Instructor in the Harris laboratory was recently awarded a R03 research grant, funded by the National Institute on Aging (NIA). This two-year, $200,000 grant is entitled “Use of super-resolution microscopy to visualize the interaction between Alzheimer therapeutic antibodies and Aβ aggregates”. This project aims to identify the molecular mechanism of action of several different clinical antibodies by localizing them on single Aβ aggregates. This information will contribute significantly to understanding the basic biology of amyloid and its role in Alzheimer’s disease, and will provide a fuller picture of molecular mechanisms responsible for detoxifying oligomers and enhancing their clearance.
Ladan Amin an Instructor in the Harris laboratory was recently awarded a R03 research grant, funded by the National Institute on Aging (NIA). This two-year, $200,000 grant is entitled “Use of super-resolution microscopy to visualize the interaction between Alzheimer therapeutic antibodies and Aβ aggregates”. This project aims to identify the molecular mechanism of action of several different clinical antibodies by localizing them on single Aβ aggregates. This information will contribute significantly to understanding the basic biology of amyloid and its role in Alzheimer’s disease, and will provide a fuller picture of molecular mechanisms responsible for detoxifying oligomers and enhancing their clearance.
Franzblau Travel Award Recipients
The Dr. and Mrs. Franzblau Travel Award Committee is pleased to announce the winners of this year’s Myrna and Carl Franzblau Student Travel Awards. We had many deserving applications this year and were able to provide awards to four PhD students and four Postdoctoral Associates within the Department. The recipients and their advisors are listed below:
- Joe Kern (Varelas)
- Addie Matschulat (Varelas)
- Jarrod Moore (Emili)
- Matt Lawton (Emili)
- Tara Liyanage, Ph.D. (Costello)
- Nachen Yang, Ph.D. (Lau)
- Robert Mercer, Ph.D. (Harris)
- Zheng Zhu, Ph.D. (Lau)
These awards are made possible by the generosity of Dr. and Ms. Franzblau and will provide funds to defer the costs of travel for these trainees to present their research at a national/international scientific conference. This year’s award selection committee was comprised of members of the Biochemistry Department Dr. and Mrs. Franzblau Travel Award Committee (Barbara Schreiber (chair), Alla Grishok, and Steve Farmer). The Department thanks the selection committee for their careful consideration of all the applicants.
Congratulations!
Cohesin organizes the genome
A new review by first author Carlos Perea-Resa in the Blower lab was recently published in Trend in Cell Biology (Cohesin: Behind Dynamic Genome Topology and Gene Expression Reprogramming)
In spite of sharing the same DNA molecules, cells in our body show different shape and function as a consequence of differential gen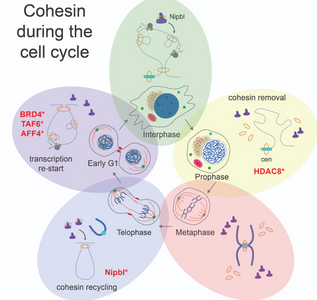 e expression. Studying how cells organize and read the instructions contained in the DNA is essential to fully understand organism development and the etiology of developmental diseases. A growing body of studies evidence the central role of the cohesin complex in both, organizing and reading genomic information. In this review published at Trends in Cell Biology, this article revises the most recent literature about how cohesin regulates gene expression. The article pays special attention to the role of cohesin in cell shifts across different stages of the cell cycle and during cell fate determination events in development. They also propose a scenario to explain the molecular etiology of Cornelia de Lange Syndrome, a developmental disease caused by cohesin disfunction on gene expression regulation.
e expression. Studying how cells organize and read the instructions contained in the DNA is essential to fully understand organism development and the etiology of developmental diseases. A growing body of studies evidence the central role of the cohesin complex in both, organizing and reading genomic information. In this review published at Trends in Cell Biology, this article revises the most recent literature about how cohesin regulates gene expression. The article pays special attention to the role of cohesin in cell shifts across different stages of the cell cycle and during cell fate determination events in development. They also propose a scenario to explain the molecular etiology of Cornelia de Lange Syndrome, a developmental disease caused by cohesin disfunction on gene expression regulation.