The Marintchev Lab
Research
Molecular mechanisms of eukaryotic translation initiation and its regulation
Control of protein synthesis (translation) is vital for cell proliferation and differentiation. Initiation of translation is typically rate-limiting and is the main target of regulation. In cancer cells, multiple components of the translation initiation machinery are up-regulated in response to the demand for high rates of protein synthesis. The potential of using inhibitors of translation initiation for anti-cancer therapy was demonstrated in recent years and currently presents an active area of research.
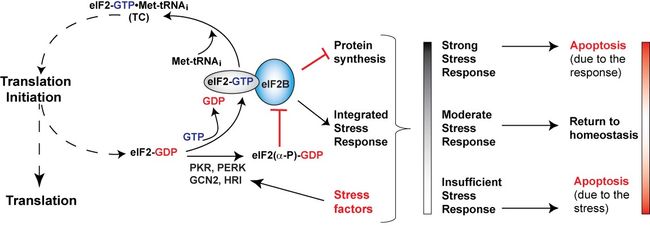
Translation initiation is the process of locating the correct translation start codon on the mRNA and the assembly of an active ribosome, ready for translation. It requires a number of eukaryotic translation initiation factors (eIFs) and consists of several steps: initiation complex assembly; binding to mRNA; scanning; start codon recognition; and finally joining of the large ribosomal subunit to form a ribosome with a bound initiator Met-tRNAi ready to translate the mRNA. The initiator Met-tRNAi is delivered to the ribosome as a complex with eIF2-GTP. One eIF2-GTP:Met-tRNAi complex is “consumed” in every translation initiation cycle, with release of eIF2-GDP and deacylated initiator tRNAi.
Regeneration of the eIF2-GTP:Met-tRNAi complex from eIF2-GDP and Met-tRNAi is catalyzed by the nucleotide exchange factor (GEF) eIF2B, a major target of regulation in response to a number of stress factors, in a process known as the integrated stress response (ISR). Dysregulated ISR is a causative factor in Alzheimer’s Disease and other neurodegenerative disorders. Our work is focused on studying the architecture of the translation initiation complexes, the molecular mechanisms of key steps in the process, and their regulation. An area of particular interest is the coordination between translation initiation and ISR.
PhD Student and Postdoctoral Fellow Positions Available
If you are interested in joining our group, please inquire at amarint@bu.edu.
Publications
For an up-to-date list of publications on PubMed please click here:
1. Marintchev, A., Ito, T. eIF2B and the Integrated Stress Response: A Structural and Mechanistic View. Biochemistry. 59(13):1299-1308 (2020).
2. Lin, K.Y., Nag, N., Pestova, T.V., Marintchev, A. Human eIF5 and eIF1A compete for binding to eIF5B. Biochemistry. 57(40):5910-20 (2018).
3. Yu, J., Marintchev, A. Comparative sequence and structure analysis of eIF1A and eIF1AD. BMC Structural Biology. 18(1):11 (2018)
4. Bogorad, A.M, Lin, K.Y, Marintchev, A. eIF2B Mechanisms of Action and Regulation: A Thermodynamic View. Biochemistry. 57(9):1426-35 (2018).
5. Bogorad, A.M, Lin, K.Y, Marintchev, A. Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res ,45(20):11962-11979 (2017).
6. Nag, N., Lin, K.Y., Edmonds, K.A., Yu, J., Nadkarni, D., Marintcheva, B., & Marintchev, A. eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res, 44(15),7441-56 (2016).
7. Bogorad, A. M., Xia, B., Sandor, D. G., Mamonov, A. B., Cafarella, T. R., Jehle, S., Vajda, S., Kozakov, D., Marintchev, A. Insights into the Architecture of the eIF2Bα/β/δ Regulatory Subcomplex. Biochemistry, 53 (21), pp 3432–45 (2014).
8. Marintchev, A. Roles of helicases in translation initiation: A mechanistic view. Biochim Biophys Acta, 1829(8), 799-809 (2013).
9. Marintchev, A. Methods for studying the interactions of translation factors with the ribosome. In: Dinman JD (Ed.) Biophysical approaches to translational control of gene expression. Springer, New York, NY, pp 83-101 (2013).
10. Marintchev, A. (Ed.) Fidelity and Quality Control in Gene Expression, Advances in Protein Chemistry and Structural Biology, 86. Elsevier (2012).
11. Luna, R. E., Arthanari, H., Hiraishi, H., Nanda, J., Martin-Marcos, P., Marcus, M. A., Akabayov, B., Milbradt, A. G., Luna, L. E., Seo, H-C., Hyberts, S. G., Fahmy, A., Reibarkh. M., Miles, D., Hagner, P., O’Day, E. M., Yi, T., Marintchev, A., Hinnebusch, A. G., Lorsch, J. R., Asano, K., Wagner, G. The C-Terminal Domain of Eukaryotic Initiation Factor 5 Promotes Start Codon Recognition by Its Dynamic Interplay with eIF1 and eIF2β. Cell Reports, 1(6), 689-702. (2012).
12. Abaeva, I. S., Marintchev, A., Pisareva, V. P., Hellen, C. U. T. & Pestova, T. V. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J, 30(1), 115-29. (2011).
13. Yu, Y., Abaeva, I. S., Marintchev, A., Pestova, T. V., Hellen, C. U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. NAR, 39(11), 4851-65. (2011).
14. Yu, Y., Marintchev, A., Kolupaeva, V. G., Unbehaun, A., Veryasova, T., Lai, S. C., Hong, P., Wagner, G., Hellen, C. U., Pestova, T.V. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. NAR, 37(15), 5167-82. (2009).
15. Marintchev, A., Edmonds, K. A., Marintcheva, B., Hendrickson, E., Oberer, M., Suzuki, C., Herdy, B., Sonenberg, N., Wagner, G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell, 136(3), 447-60. (2009).
16. Suzuki, C., Garces R. G., Edmonds, K. A., Hiller, S., Hyberts, S. G., Marintchev, A. & Wagner, G. The tumor suppressor PDCD4 inhibits translation initiation by binding to eIF4A using both its MA3 domains. PNAS, 105(9), 3274-9. (2008)
17. Lindqvist, L., Oberer, M., Reibarkh, M., Cencic, R., Bordeleau, M-E., Marintchev, A., Tanaka, J., Fagotto, F., Altmann, M., Wagner, G. & Pelletier, J. Selective Pharmacological Targeting of a DEAD box RNA helicase, PLoS ONE, 3(2): e1583, (2008).
18. Marintcheva, B., Marintchev, A., Wagner, G. & Richardson C. C. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. PNAS, 105(6), 1855-60. (2008)
19. Unbehaun, A., Marinchev, A., Lomakin, I. B., Didenko, T., Wagner, G., Hellen, C. U. T., & Pestova, T. V. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing, EMBO J, 26, 3109-3123, (2007).
20. Marintchev, A., Frueh, D., & Wagner, G. NMR methods for studying protein-protein interactions involved in translation initiation. Meth Enz, 430, 283-331, (2007).
21. Gelev, V., Aktas, H., Marintchev, A., Ito, T., Frueh, D., Hemond, M., Rovnyak, D., Debus, M., Hyberts, S. G., Usheva, A., Halperin, H., & Wagner, G. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. JMB, 364(3), 352-63, (2006).
22. Yamamoto, Y., Singh, C. R., Marintchev, A., Hall, N. S., Hannig, E. M., Wagner, G., & Asano, K. The C-terminal HEAT domain of eIF5 mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3 and eIF4G. PNAS 102(45), 16164-9 (2005).
23. Marintchev, A. & Wagner, G. eIF4G and CBP80 share a common origin and similar domain organization – implications for the eIF4G structure and function. Biochemistry 44(37), 12265-72 (2005).
24. Oberer, M., Marintchev, A. & Wagner, G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19(18), 2212-23 (2005).
25. Marintchev, A. & Wagner, G. Translation initiation: structures, mechanisms and evolution, Q Rev Biophys 37(3-4), 197-284 (2004).
26. Ito,T., Marintchev, A. & Wagner, G. Solution Structure of Human Initiation Factor eEF1B Reveals Homology to the Elongation Factor eEF1B. Structure (Camb) 12(9), 1693-704 (2004).
27. Marintchev, A., Kolupaeva, V. G., Pestova, T. V. & Wagner, G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. PNAS 100(4), 1535-40 (2003).
28. Lomakin, I. B., Kolupaeva, V. G., Marintchev, A., Wagner, G. & Pestova, T. V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17(22): 2786-97 (2003).
29. Marintchev, A., Gryk, M. R. & Mullen, G. P. Site-directed mutagenesis analysis of the structural interaction of the single-strand-break repair protein, X-ray cross-complementing group 1, with DNA polymerase β. NAR 31(2), 580-8 (2003).
30. Gryk, M. R., Marintchev, A. & Mullen, G. P. Mapping of the interaction interface of DNA polymerase β with XRCC1. Structure (Camb) 10(12), 1709-20 (2002).
31. Maciejewski, M. W., Shin, R., Pan, B., Marintchev, A., Denninger, A., Mullen, M. A., Chen, K., Gryk, M. R. & Mullen, G. P. Solution Structure of a Viral DNA Repair Polymerase. Nature Struct Biol 8(11), 936-41 (2001).
32. Pan, B., Maciejewski, M. W., Marintchev, A. & Mullen, G. P. Solution structure of the catalytic domain of γδ-resolvase. Implications for the mechanism of catalysis. JMB 310(5), 1089-107 (2001).
33. Wu, H., Maciejewski, M. W., Marintchev, A., Benashski, S. E., Mullen, G. P. & King, S. M. Solution structure of a dynein motor domain-associated light chain. Nature Struct Biol 7(7), 575-9 (2000).
34. Marintchev, A., Robertson, A., Dimitriadis, E. K., Prasad, R., Wilson, S. H., & Mullen, G. P. Domain specific interaction in the XRCC1-DNA polymerase β complex. NAR 28(10), 2049-59 (2000).
35. Marintchev, A., Mullen, M. A., Maciejewski, M. W., Pan, B., Gryk, M. R. & Mullen, G. P. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nature Struct Biol 6(9), 884-93 (1999).
36. Marintchev, A., Maciejewski, M. W. & Mullen, G. P. 1H, 15N, and 13C resonance assignments for the N-terminal 20 kDa domain of the DNA single-strand break repair protein XRCC1. J Biomol NMR 13(4), 393-4 (1999).
37. Marintchev, A., Mirtcheva, J., Sidjimov, A. & Haimova, M. Induction of lymphoproliferative popliteal lymph node reaction by hydantoin derivatives: structure-activity relationships. Int J Immunopharmacol 17(12), 981-4 (1995).
Links
BU Profile
ResearchGate
LinkedIn
Contact Us
Department of Pharmacology, Physiology & Biophysics
Chobanian & Avedisian School of Medicine
700 Albany Street, W336A
Boston, MA 02118-2526
Phone: (617) 358-8486
e-mail: amarint@bu.edu