The Bullitt Lab
Working Together to Promote an Inclusive Community
I SUPPORT EQUITY AND INCLUSION, AND THE BLACK LIVES MATTER MOVEMENT
Bullitt Lab Research
Protein Structure Facilitates Function
Using Electron Microscopy and Quantitative Image Analysis to see how Macromolecular Assemblies Work
Structural studies of biological macromolecular assemblies are providing a deeper understanding of cellular function. In our laboratory, we utilize electron microscopy and image reconstruction to investigate questions about:
- how adhesion pili aid pathogenic bacterial survival
- how viruses impact cellular processes
- how the type III secretion system is assembled, leading to secretion of toxins
BACTERIAL ADHESION PILI
YouTube: the role of basic research in developing a vaccine against Traveler’s Diarrhea: Vaccines from basic research
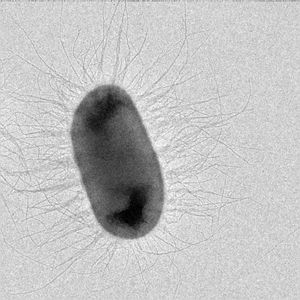
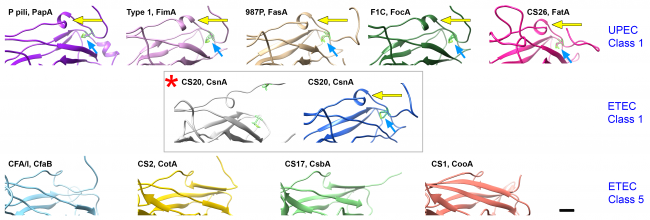
Fibers on the surfaces of many pathogenic bacteria can overextend like a toy Slinky, whereas homologous proteins on bacteria that cause pneumonia, ear infections, and meningitis assemble into a 3-stranded rope-like fibers (blue). In collaboration with Dr. Magnus Andersson’s lab at Umeå University in Umeå Sweden and Dr. Joseph Baker’s lab at the College of New Jersey we have shown that the forces required for unwinding pili (also called ‘fimbriae’) are specific for the pilus-type, and thus vary with the microenvironment expected to be encountered. For example, CFA/I pili on diarrhea-causing intestinal bacteria unwind at less than one-third the force required to unwind P-pili expressed on urinary tract infection bacteria.
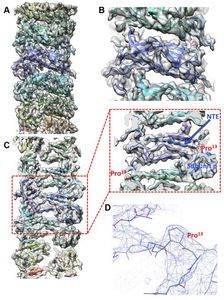
Our cryo-electron microscopy (cryo-EM) results now show the structure of CFA/I pili at 3.5 A resolution, previously published at 4.3 A resolution in
IUCrJ (2019) 6, 815–821,
https://doi.org/10.1107/S2052252519007966
As more bacteria become resistant to antibiotics, it is essential to develop novel approaches for prevention and treatment of infection. Structural studies are vital for discovering clues to how bacteria bind, and how they remain attached while the host is trying to remove them. We are examining the architecture of bacterial adhesion pili and investigating small molecules that disrupt their assembly and/or function. We see that pilin subunits must rotate to form a helical filament, after exiting linearly from the bacterial surface.
Selected Publications
Bullitt, E., L. Makowski (1995). Structural polymorphism of bacterial adhesion pili. Nature 373:164-167. PMID: 7816100
Brown JW, Badahdah A, Iticovici M, Vickers TJ, Alvarado DM, Helmerhorst EJ, Oppenheim FG, Mills JC, Ciorba MA, Fleckenstein JM, Bullitt E (2018). A Role for Salivary Peptides in the Innate Defense Against Enterotoxigenic Escherichia coli. J Infect Dis. 2018 217:1435-1441. doi: 10.1093/infdis/jiy032. PMID 29528423. PMC5894089
Baker JL, Dahlberg T, Bullitt E, Andersson M (2021). Impact of an alpha helix and a cysteine-cysteine disulfide bond on the resistance of bacterial adhesion pili to stress. Proc Natl Acad Sci U S A. 118:e2023595118. doi: 10.1073/pnas.2023595118. PMID 34011607. PMC8166124
CHANGES TO CELLS DURING VIRUS REPLICATION
Extracellular vesicles secreted from virus-infected cells contain components necessary to begin virus replication quickly, including viral RNA and proteins as well as host proteins.
Yang JE, Rossignol ED, Chang D, Zaia J, Forrester I, Raja K, Winbigler H, Nicastro D, Jackson WT, Bullitt E (2020). Complexity and ultrastructure of infectious extracellular vesicles from cells infected by non-enveloped virus. Sci Rep. 10:7939. doi: 10.1038/s41598-020-64531-1. PMID 32409751. PMCID: PMC7224179
We are using our expertise from studies on poliovirus replication, we have multiple collaborators at BUSM for studies on Zika Virus, SARS-CoV2, and filoviruses.
Rossignol ED, Peters KN, Connor JH, Bullitt E (2017). Zika virus induced cellular remodelling. Cell Microbiol. 19:10.1111/cmi.12740. doi: 10.1111/cmi.12740. PMID 28318141. PMC5561415
In collaboration with Dr. Karla Kirkegaard’s lab at Stanford University we have shown that “Zombie” 3Dpol protein (dead active site) can support assembly of oligomers and restore replication when the wild type protein concentration is too low to do so.
Spagnolo, J.F., E. Rossignol, E. Bullitt, K. Kirkegaard (2010). Enzymatic and non-enzymatic functions of viral RNA-dependent RNA polymerases within oligomeric arrays. RNA 16:382-393. PMCID: PMC2811667
TYPE III SECRETION
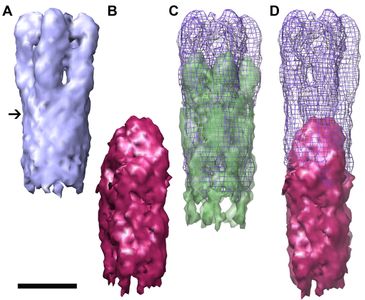
Only 10 bacteria (or fewer!) are needed for Shigella flexneri to cause dysentery (bloody diarrhea). This disease is initiated via the type III secretion system, which is used to secrete both toxins and bacterial effectors that alter normal host cell functions to promote bacterial growth and spread.
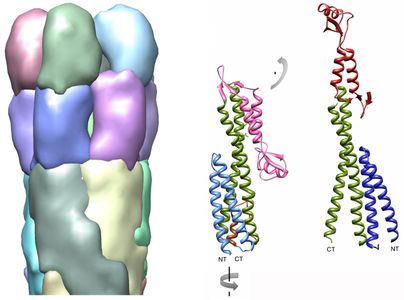
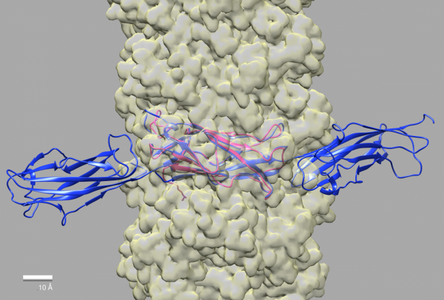
To understand (and disrupt) the process of infection, we are examining assembly intermediates of the T3SS syringe-like needle and its tip structure. Through our collaborations with the Picking lab at Oklahoma State University Stillwater and the Geisbrecht lab at the University of Missouri Kansas City we have shown that IpaD, the first protein that localizes to the needle tip, is present as a pentamer, and undergoes a dramatic conformational change as compared to the structure that has been solved by x-ray crystallography.
The structures of three-dimensional reconstructions of immature and nascent needle tips show clearly IpaD as an elongated pentameric structure.
This new result demonstrates that the distal domain of IpaD flips up, as compared to the structure solved by X-ray crystallography (pdb 2j0o).
Diagram to the right shows the conformational change of the distal domain that is needed to fit the crystal structure of IpaD into our density map of 3-dimensional reconstructions from electron microscopy data.
Epler, C.R., N.E. Dickenson, E. Bullitt*, W.L. Picking* (2012). Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J. Mol. Biol. 420:29-39. PMID: 22480614 *corresponding authors
Links:
Faculty Profile
ORCID
MyNCBI
PubMed Citations
Selected Media Coverage:
Diarrheal Disease Protection May Literally Be within Spitting Distance
Contact Us
Esther Bullitt, Ph.D.
Department of Pharmacology, Physiology & Biophysics
Chobanian & Avedisian School of Medicine
700 Albany Street
Boston MA 02118-2526
Phone: (617) 358-8464
e-mail: bullitt@bu.edu