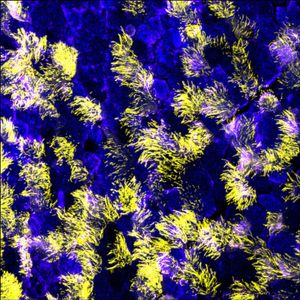
Each time a cell divides the correct number of chromosomes must be segregated to each daughter cell. Failures in chromosome segregation lead to an incorrect chromosome content (aneuploidy), which is highly correlated with cancer and is a leading cause of birth defects and developmental diseases in humans. Accurate chromosome segregation requires a dramatic transformation of the genome, which impacts all aspects of chromatin structure and RNA metabolism.
The long-term goal of my laboratory is to understand proteins that link RNA to DNA and chromokinesins contribute to chromatin structure and DNA repair in interphase and accurate chromosome segregation during mitosis. We are currently focused on: 1) how transcription is silenced during mitosis and restarted following mitosis, 2) how non-coding RNAs are removed from chromatin during mitosis, 3) how the chromokinesin Kif4a contributes to chromosome structure and DNA repair.
We are a multidisciplinary lab that uses cell biology, biochemistry, and genomic approaches coupled with CRISPR genome engineering. Our primary experimental systems is human cell culture.
In addition to research, I teach many classes for graduate students and have initiated a computational biology module within the FiBS classes. I am also the director of the Biochemistry and Cell Biology graduate program.
Laboratory Members
- Alice Ma – Undergraduate Student
- Jessica Daniel – Undergraduate Student
- Andrea King – Undergraduate Student
- Madeline Prechtl – Undergraduate Student
Representative Publications
Complete publication list at BU Profiles
-
Watanabe R, Perea-Resa C, Blower MD. Robust CENP-A incorporation in human cells is independent of transcription and cohesin components. Mol Biol Cell. 2025 Nov 01; 36(11):br30. PMID: 40961235; PMCID: PMC12562020; DOI: 10.1091/mbc.E25-05-0214;
-
Nakos K, Wijeratne S, Blower M, Subramanian R. A minimum module for positioning the Chromosomal Passenger Complex at the cell center for cytokinesis. bioRxiv. 2025 Sep 02. PMID: 40950017; PMCID: PMC12424632; DOI: 10.1101/2025.09.02.673761;
-
Sharp JA, Sparago E, Thomas R, Alimenti K, Wang W, Blower MD. Role of the SAF-A/HNRNPU SAP domain in X chromosome inactivation, nuclear dynamics, transcription, splicing, and cell proliferation. PLoS Genet. 2025 Jun; 21(6):e1011719. PMID: 40493679; PMCID: PMC12176297; DOI: 10.1371/journal.pgen.1011719
-
Watanabe R, Perea-Resa C, Blower MD. Robust CENP-A incorporation in human cells is independent of transcription and cohesin components. bioRxiv. 2025 May 01. PMID: 40654925; PMCID: PMC12247887; DOI: 10.1101/2025.04.29.651290;
-
Sharp JA, Sparago E, Thomas R, Alimenti K, Wang W, Blower MD. Role of the SAF-A SAP domain in X inactivation, transcription, splicing, and cell proliferation. bioRxiv. 2024 Sep 10. PMID: 39314300; PMCID: PMC11419091; DOI: 10.1101/2024.09.09.612041;
-
Park S, Dahn R, Kurt E, Presle A, VanDenHeuvel K, Moravec C, Jambhekar A, Olukoga O, Shepherd J, Echard A, Blower M, Skop AR. The mammalian midbody and midbody remnant are assembly sites for RNA and localized translation. Dev Cell. 2023 Oct 09; 58(19):1917-1932.e6. PMID: 37552987; PMCID: PMC10592306; DOI: 10.1016/j.devcel.2023.07.009;
-
Blower MD, Wang W, Sharp JA. Differential nuclear import regulates nuclear RNA inheritance following mitosis. Mol Biol Cell. 2023 Apr 01; 34(4):ar32. PMID: 36790906; PMCID: PMC10092649; DOI: 10.1091/mbc.E23-01-0004;
- Perea-Resa C, Wattendorf L, Marzouk S, Blower MD. Cohesin: Behind Dynamic Genome Topology and Gene Expression Reprogramming. Trends Cell Biol. 2021 Mar 22:S0962-8924(21)00051-9. doi: 10.1016/j.tcb.2021.03.005. PMID: 33766521.
-
Hekman RM, Hume AJ, Goel RK, Abo KM, Huang J, Blum BC, Werder RB, Suder EL, Paul I, Phanse S, Youssef A, Alysandratos KD, Padhorny D, Ojha S, Mora-Martin A, Kretov D, Ash PEA, Verma M, Zhao J, Patten JJ, Villacorta-Martin C, Bolzan D, Perea-Resa C, Bullitt E, Hinds A, Tilston-Lunel A, Varelas X, Farhangmehr S, Braunschweig U, Kwan JH, McComb M, Basu A, Saeed M, Perissi V, Burks EJ, Layne MD, Connor JH, Davey R, Cheng JX, Wolozin BL, Blencowe BJ, Wuchty S, Lyons SM, Kozakov D, Cifuentes D, Blower M, Kotton DN, Wilson AA, Mühlberger E, Emili A. Actionable Cytopathogenic Host Responses of Human Alveolar Type 2 Cells to SARS-CoV-2. Mol Cell. 2021 Jan 07; 81(1):212. PMID: 33417854
-
Ardehali MB, Damle M, Perea-Resa C, Blower MD, Kingston RE. Elongin A associates with actively transcribed genes and modulates enhancer RNA levels with limited impact on transcription elongation rate in vivo. J Biol Chem. 2020 Dec 17. PMID: 33334895
- Perea-Resa, C., Bury, L., Cheeseman, I.C., and Blower MD. Cohesin removal reprograms gene expression upon mitotic entry. Mol Cell. 2020 Apr 2;78(1):127-140.e7. PMID: 32035037
- Yang, F. Wang, W. Cetinbas, M., Sadreyev, R.I., and Blower, MD. Genome-wide analysis identifies cis-acting elements regulating mRNA polyadenylation and translation during vertebrate oocyte maturation. RNA. 2020 Mar;26(3):324-344. PMID: 31896558
- Sharp JA, Perea-Resa C, Wang W, Blower MD. Cell division requires RNA eviction from condensing chromosomes. J Cell Biol. 2020 Nov 02; 219(11). PMID: 33053167
- Toombs JA, Sytnikova YA, Chirn GW, Ang I, Lau NC, Blower MD. Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome. RNA. 2016 Dec 28. pii: rna.058859.116. doi: 10.1261/rna.058859.116. PMID: 28031481.
- Blower, M.D. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep. 2016. May 24. 15(8):1624-1633 PMID: 27184843
- Jambhekar A, Emerman AB, Schweidenback CT, Blower MD. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 2014 Jun 26;9(6):e100748. PMID: 24968351
- Sharp, J., Plant, J., Ohsumi, T., Borowsky, M., Blower, MD. Functional analysis of the microtubule-interacting transcriptome. Mol Biol Cell. 2011. Nov;22(22):4312-23. PMID: 21937723Complete list available on BU Profiles