Congratulations to Stefan Isaac; one of the selected 2025 Toffler Scholars!
Congratulations to Stefan Isaac who was one of the selected 2025 Toffler Scholars!

The Toffler Scholar Program supports medical investigators engaged in early-stage, innovative medical research.
See announcement here:
Karen Toffler Charitable Trust Announces Toffler Scholars - Toffler Trust
New Research from the Layne Lab: mechanisms of adipose tissue fibrosis
A new study from the Layne laboratory identified a function for the transcription factor FOXS1 in regulating TGFb-dependent changes in adipogenic differentiation: JBC in press.
White adipose tissue (WAT) fibrosis is a major determinant of obesity-induced dysfunction and is characterized by excessive extracellular matrix deposition and myofibroblast activation. In this study, first author Alexander Tavares and co-authors identified FOXS1, a member of the forkhead box transcription factor superfamily, as a transcriptional target of TGF-β1 signaling in primary human adipocyte stem cells. FOXS1 also attenuated the induction of several adipogenic factors and sensitized cells to the anti-adipogenic effects of TGF-β1. Furthermore, loss of endogenous FOXS1 improved adipogenic permissiveness and activated proadipogenic gene programs in progenitors, even after TGF-β1 stimulation. These results indicate that FOXS1 is a positive regulator of profibrotic TGF-β1-dependent cellular responses, orchestrating the regulation of molecular phenotypes that promote myofibroblast activation and block adipogenesis. Co-authors in this study from the Layne lab include Scott Connelly, Daryn Maksat, Jane Zheng and Nabil Rabhi from the Farmer lab.
New news out of the Saeed Lab: Researchers Discover Protein Necessary for SARS-CoV-2 to Evade the Body’s Defenses
Congratulations to Barbara Schreiber being named educator of the year
Congratulations to Dr. Barbara Schreiber who has been named the 2025 Educator of the Year: PhD. 
New research out of the Lau Lab: Researchers Discover Protein Necessary for Fruit Fly Fertility
Researchers Discover Protein Necessary for Fruit Fly Fertility
The global birthrate has been in significant decline for decades. In the U.S., couples are deciding to have children later in life. A 2022 U.S. Census data analysis of Census Bureau and National Center for Health Statistics data, reveals that fertility rates for women age 20-24 declined by 43% during the period from 1990 to 2013. But the numbers of women age 35-39 giving birth increased by 67%, and for women between 40- 44 that increase was nearly 139%.
Women who decide to have children in middle age depend on sperm and egg resiliency. Part of germ cell (also known as egg cells and sperm cells) resilience depends on a functional piRNA pathway to protect germ cell genomes decades after puberty, which is when in humans, the Piwi pathway is activated as well as the expression of transposon RNAs—mobile DNA sequences that can move around a genome.

Researchers from Boston University Chobanian & Avedisian School of Medicine have found a new role for the transcription factor (proteins that regulate the transcription, or copying, of genes). In the fruit fly, this transcription factor, named Traffic Jam, activates a non-coding piRNA gene named Flamenco to promote female fruit fly (drosophila) fertility. The discovery solves the 30-year-old mystery of how Flamenco gets activated to protect fruit fly ovaries from a series of genetic parasites called retroviral transposons, and may one day help with infertility issues in humans.
“The discovery of Traffic Jam’s function in flies will help us look into humans with infertility symptoms and see if those patients lacking functional sperm may have defects in these Piwi genes and transcription factors,” explains corresponding author Nelson Lau, PhD, associate professor of biochemistry and director of the BU Genome Science Institute.

Lau and his colleagues first conducted a series of luciferase-reporter assays that measure gene activity and biological responses in 2017, discovering important new regulatory sequences in the Flamenco locus. They confirmed the biological importance of those Flamenco DNA sequences by generating new fruit fly mutants with CRISPR genome editing. They then carried out proteomics experiments and made the first discovery of Traffic Jam binding to Flamenco DNA sequences. Lastly, they conducted RNA interference knockdowns and chromatin immunoprecipitation sequencing studies of Traffic Jam in fruit fly ovary cells to confirm this new genetic interaction.
They found that Traffic Jam driving the production of Flamenco piRNAs bound by Piwi proteins, allowed fruit flies to protect their germline genome and produce fertile eggs and offspring. They also found that the retroviral transposons are also activated by Traffic Jam, hijacking this host factor. This finding is surprising because it demonstrates the on-going war between the animal’s genetic immune system and the genetic parasites fighting for their own survival.
According to the researchers, this study investigates the fundamental battle for the protection of germline genomes that humans depend on for fertility and reproduction. “We humans are like fruit flies in that our gonads also generate piRNAs to protect our germ cells against transposons. We have our own version of the Traffic Jam gene, called MAF-B, which we can test in future studies to see if MAF-B regulated human piRNA genes to allow us to produce functional sperm,” adds Lau.
These findings appear online in the journal Cell Reports. Lau’s team also contributed to an accompanying study in the same issue of Cell Reports by researchers in France and the United Kingdom.
Congratulations to Jean Spencer on her recent promotion to Assistant Professor
Congratulations to Jean Spencer on her recent promotion to Assistant Professor
Congratulations to Mohsan Saeed on his recent promotion to Associate Professor of Biochemistry & Cell Biology!
Congratulations to Mohsan Saeed on his recent promotion to Associate Professor of Biochemistry & Cell Biology!

Check out this article on The Brink featuring Dr. Mohsan Saeed’s research. “West Nile, Zika, Coronavirus: This BU Researcher is Taking on Some of the World’s Most Serious Viruses—And He Plans to Win”

West Nile, Zika, Coronavirus: This BU Researcher is Taking on Some of the World’s Most Serious Viruses—And He Plans to Win
New publication out of the Perrissi Lab “Inhibition of K63 ubiquitination by G-Protein Pathway Suppressor 2 (GPS2) regulates mitochondria-associated translation”
The Perissi Lab has recently published in Pharmacological Research a paper titled “Inhibition of K63 ubiquitination by G-Protein Pathway Suppressor 2 (GPS2) regulates mitochondria-associated translation”.
The paper reports the characterization of a novel strategy for regulating mitochondria-associated translation based on the mitochondria retrograde factor GPS2 modulating non-proteolytic ubiquitination of pmitochondria-associated translation factors. Removal of GPS2, either by genomic deletion or via stress-induced translocation to the nucleus, licenses K63 ubiquitination of poly-A binding protein PABPC1 and other factors, favoring translation initiation and promoting the expression of an anti-oxidant stress response program.
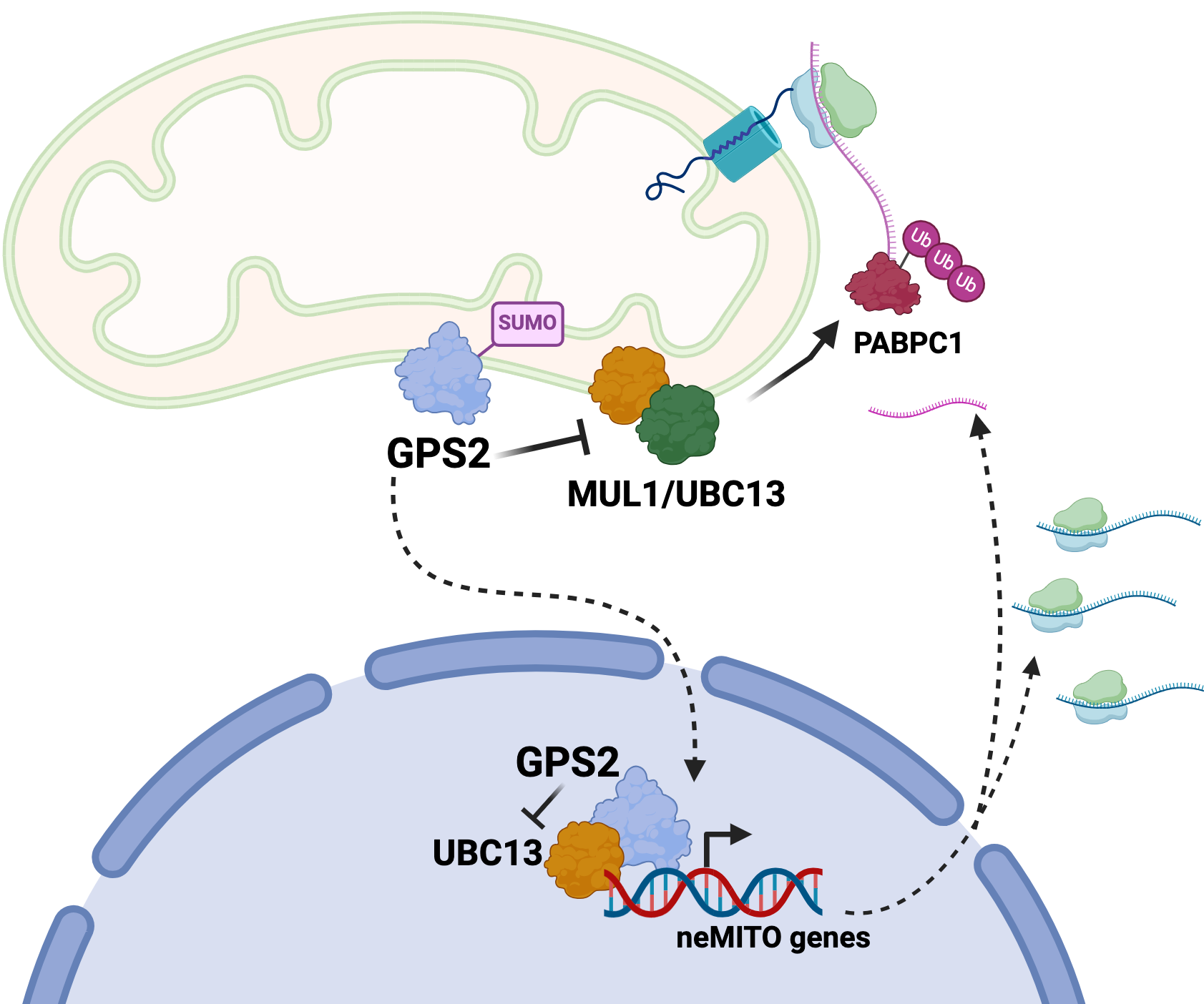
Translation of selected nuclear-encoded mitochondrial mRNAs on mitochondria-associated ribosomes provides a unique opportunity for regulating protein translation at single organelle level and thus for coordinating gene expression with mitochondrial health. These findings, together with the previous characterization of nuclear GPS2 as an essential cofactor for the expression of nuclear-encoded mitochondrial genes, reveal that GPS2-mediated inhibition of K63 ubiquitination provides a unifying strategy for modulating the expression of mitochondrial proteins through coordinated transcription and translation events.
The study was led by former postdoctoral fellow Dr. Yuan Gao, in collaboration with the Center for Network Systems Biology and the Lyons Lab.
Welcome New Faculty member – Stefan Isaac, Ph.D.
We are happy to announce that Stefan Isaac, Ph.D., recently accepted our offer to join the department as an Assistant Professor as of July 1, 2024.

Dr. Isaac is currently a postdoc with Stirling Churchman at Harvard where he has been carrying out pioneering studies of the mitochondrial genome, a subject that is poorly understood, and whose investigation poses significant experimental challenges. His work is a perfect fit for the genomic and cell biological directions of the department, and he will be able to collaborate with a number of our current faculty members.
Please join us in welcoming Stefan!
