Congratulations to David A. Harris, M.D.,Ph.D. on being awarded The Edgar Minas Housepian, MD, Professorship
Congratultions to David A. Harris on being one of 5 recipients of the Edward Avedisian Professorships.

The Edgar Minas Housepian, MD, Professorship went to David A. Harris, chair of biochemistry andcell biology since 2009. He studies molecular and cellular mechanisms underlying human neurodegenerative diseases. His work on infectious prion diseases like mad cow disease, where brain proteins fold and can result in neurodegenerative effects, has helped research into other neurodegenerative diseases, like Alzheimer’s, Parkinson’s, and Huntington’s.
“What I want to highlight here is the incredible foresight to use that endowment ($25 million of the Avedisian endowment is dedicated to research and teaching) to support basic research, which is…always at the root of great medical discoveries,” said Harris. “I am honored to be associated with a legacy that values the pursuit of knowledge and scientific excellence.”
Housepian was a renowned neurosurgeon at New York-Presbyterian Hospital and professor of neurology at Columbia University’s medical school, where he taught for 44 years.
“He was a very creative person with a long-range vision,” said his daughter, Jean Housepian. Even with a career that began in labs, then surgery, education remained his key concern, and in his retirement years, he remained an advocate for international educational affiliations for medical students.
See the whole article here.
Congratulations to Mike Blower on his recent promotion to Full Professor of Biochemistry & Cell Biology!
Congratulations to Mike Blower on his recent promotion to Full Professor of Biochemistry & Cell Biology!

The Blower lab is a multidisciplinary lab that uses cell biology, biochemistry, and genomic approaches coupled with CRISPR genome engineering. Their primary experimental systems are human cell culture and frog oocytes.
Mikel Garcia-Marcos elected Chair of Division of National Scientific Society

Mikel Garcia-Marcos has been elected as Chair of the Division of Molecular Pharmacology of the American Cancer Society of Pharmacology and Experimental Therapeutics (ASPET).
He will serve a 1-year term as Chair-Elect followed by a 1-year term as Chair. In this role, he will oversee the activities of the Executive Committee involving planning of the Society's Annual Meeting, financially supporting external scientific meetings, selection of scientific achievement awardees, and organizing other activities like online webinars and workshops.
The American Society for Pharmacology and Experimental Therapeutics (ASPET) is a 4,000 member scientific society whose members conduct basic and clinical pharmacological research and work for academia, government, large pharmaceutical companies, small biotech companies, and non-profit organizations. ASPET is a global pharmacology community that advances the science of drugs and therapeutics to accelerate the discovery of cures for disease.
Congratulations to Daniel Cifuentes on his recent promotion to Associate Professor of Biochemistry & Cell Biology!
Congratulations to Daniel Cifuentes on his recent promotion to Associate Professor of Biochemistry & Cell Biology!

Research goals in the Cifuentes laboratory aim to understand the molecular mechanisms of RNA regulation. In particular, they focus on the study of RNA-binding proteins, microRNAs, and RNA modifications to uncover their impact on small RNA biogenesis and mRNA post-transcriptional regulation.
New publication sheds light onto the molecular basis of neurotransmitter signaling
A new paper led by Alex Luebbers in the Garcia-Marcos Lab dissects the molecular basis for how GINIP, a Gαi-interacting protein in neurons involved in controlling pain and seizures, modulates GPCR responses triggered by neurotransmitters. GINIP mimics how G-protein signaling effectors bind to Gα subunits via its PHD domain to differentially scale discrete G-protein signaling branches. The paper has been published in Structure (https://www.sciencedirect.com/science/article/abs/pii/S0969212623003891?via%3Dihub), and was previously posted on bioRxiv (https://www.biorxiv.org/content/10.1101/2023.04.20.537566v1).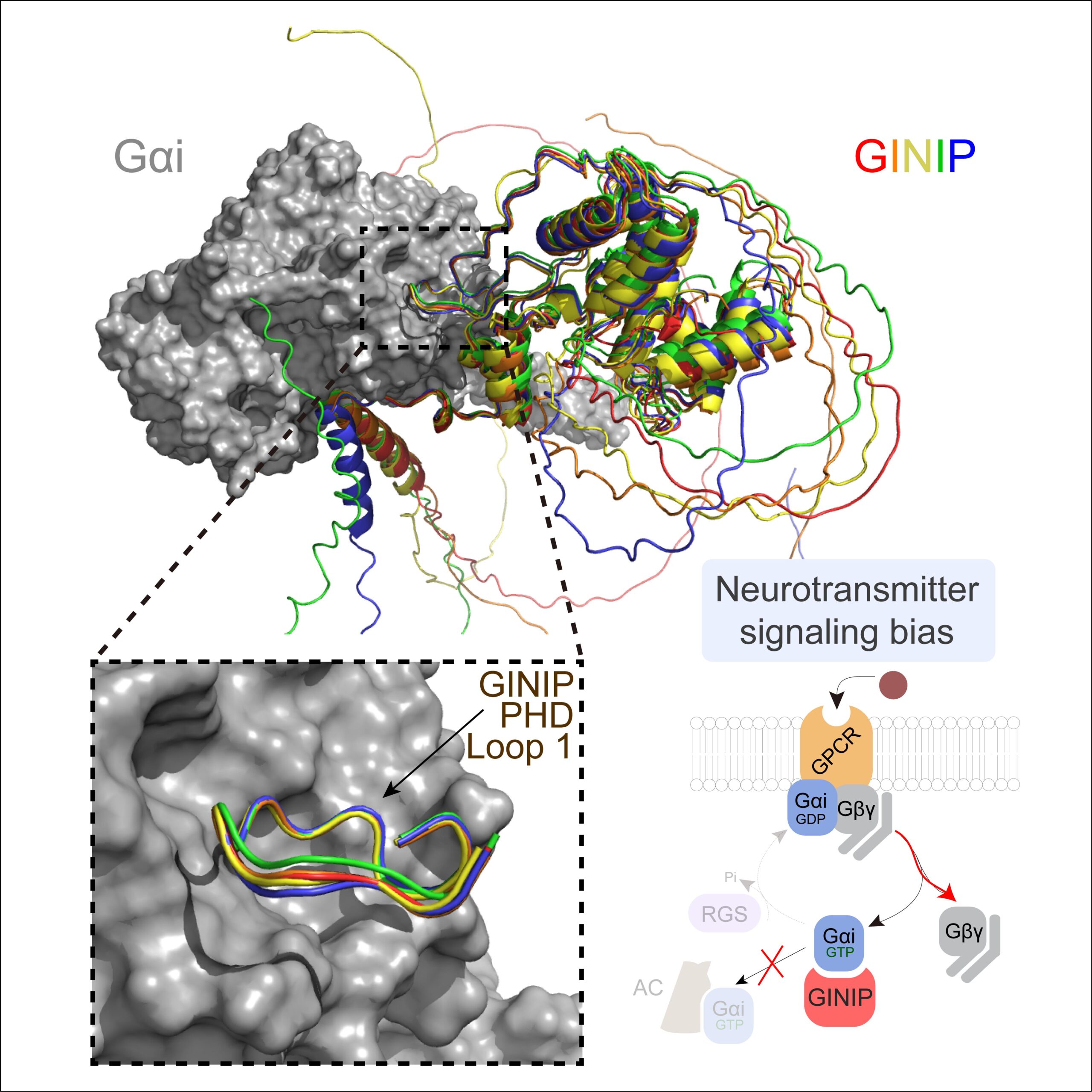
This paper builds and explands on another paper recently published in the same lab establishing that the neuronal protein GINIP shapes GPCR inhibitory neuromodulation via a unique mechanism of G-protein regulation that controls pain and seizure susceptibility (Park, Luebbers, et al Molecular Cell, 2023). However, the molecular basis of this mechanism remained ill-defined because the structural determinants of GINIP responsible for binding and regulating G proteins were not known. The newly published paper combined hydrogen-deuterium exchange mass spectrometry, computational structure predictions, biochemistry, and cell-based biophysical assays to demonstrate an effector-like binding mode of GINIP to Gαi. These findings explain the molecular basis for a post-receptor mechanism of G-protein regulation that fine-tunes inhibitory neuromodulation.
This paper features collaborations with the labs of Stephen Eyles at UMass-Amherst and of Joshua Levitz at Weill Cornell.
Mohsan Saeed, PhD, Honored with 2023 Smith Family Foundation Odyssey Award
M ohsan Saeed, PhD, Assistant Professor of Biochemistry & Cell Biology, has been selected as a recipient of the 2023 Smith Family Foundation: Odyssey Award. He is the first recipient from BU to get the Smith Odyssey Award.
ohsan Saeed, PhD, Assistant Professor of Biochemistry & Cell Biology, has been selected as a recipient of the 2023 Smith Family Foundation: Odyssey Award. He is the first recipient from BU to get the Smith Odyssey Award.
As part of this honor, Saeed will receive $400,000 to study how mosquito-borne viruses (arboviruses) overcome the immune system of mosquitoes and establish persistent infection that is then transmitted to humans. He will collaborate on this project with Joseph Zaia, PhD, professor of biochemistry & cell biology, and Fabiana Feitosa-Suntheimer, PhD, a senior research scientist at BU’s National Emerging Infectious Diseases Laboratories (NEIDL).
Mosquito-borne arboviruses claim over one million human lives each year and are considered a global health priority due to frequent resurgence of activity and unprecedented geographical expansion in recent decades. In the absence of vaccines and targeted treatments, the design of strategies to control arboviruses at the mosquito level is imperative.
According to Saeed, his project is based on the premise that an in-depth understanding of mechanisms by which arboviruses establish lifelong infection in mosquitoes can inspire the design of powerful approaches to reduce viral transmission to humans. He will use an advanced technique, which he recently developed, to investigate the molecular details as to how arboviruses disarm mosquitoes’ antiviral defense systems and establish persistent infection.
“These studies will open up new lines of investigation into viral persistence and mosquito biology and facilitate the design of transgenic mosquitoes unable to harbor and transmit infections. It will also accelerate a discovery pipeline that can then be extended to other insect-borne pathogens such as plasmodium and borrelia,” said Saeed, who also is an investigator at the NEIDL.
Saeed received his MPhil in microbiology from Quaid-e-Azam University, Pakistan, where he studied the molecular epidemiology of polio-like viruses in patients suffering from paralysis. He then joined the University of Tokyo, receiving his PhD in Pathology, Immunology and Microbiology. During his doctoral studies, he developed novel cell culture systems for the study of hepatitis C virus (HCV) and investigated various aspects of this virus in diverse in vitro and in vivo settings.
Saeed then entered the laboratory of Nobel laureate Charles M. Rice, PhD, at The Rockefeller University, New York, for his postdoctoral training. Although his research in the Rice Lab mainly focused on HCV, he also gained expertise with a number of other positive-strand RNA viruses, including enteroviruses, flaviviruses and alphaviruses. In addition, Saeed developed a novel viral degradomics technique that allows an unbiased identification of cellular proteins cleaved during viral infections.
Saeed joined BU in 2019; his lab explores the role of viral and host proteases in disease mechanisms of positive-strand RNA viruses at the NEIDL. In early 2021 when COVID-19 was declared a global pandemic, his lab pivoted to SARS-CoV-2 research and has since made contributions to the molecular understanding of how SARS-CoV-2 establishes infection in various tissues and interacts with the human innate and adaptive immune systems.
The Richard and Susan Smith Family Foundation created the award in 2017 to fuel creativity and innovation in junior investigators in the basic sciences. It supports the pursuit of high impact ideas to generate breakthroughs and drive new directions in biomedical research. The award funds high-risk, high-reward pilot projects solicited from the brightest junior faculty in the region.
Biochemistry & Cell Biology 2023 Retreat to Thompsons Island
On September 22. 2023 the Department of Biochemistry & Cell Biology ventured out to Thompsons Island in the Boston Harbor for its annual retreat. We are a large department, and the retreat provides an opportunity to bring people together to learn about the work taking places across different laboratories and specialties, as well as meet people from the different groups.

The agenda included a 'state of the department' delivered by department chair Dr. David Harris, presentations by several PIs within the department. At the end of the day we held a Poster Session where trianees in the labs were able to present their work and we voted on winners!

Biochemistry & Cell Biology Faculty Position Available – Assistant Professor
The Department of Biochemistry and Cell Biology at the Boston University Chobanian and Avedisian School of Medicine (https://www.bumc.bu.edu/biochemcellbio/) invites application for a faculty position at the Assistant Professor level. The position comes with competitive salary, generous start-up support, and ample lab and office space.
We are seeking scientists with a demonstrated record of productivity and scientific excellence, addressing important and challenging problems across a broad range of research areas, including, but not limited to, cell biology, systems biology, epigenetics and chromatin biology, RNA biology, and metabolism. Candidates must have a Ph.D. and/or M.D. degree, with relevant postdoctoral experience, and plans to develop a creative and independent research program. We especially welcome applications from members of all groups underrepresented in STEM.
The successful candidate will join a welcoming and vibrant Department that in the last decade has enjoyed a major transformation in its personnel and infrastructure, including hiring new faculty members at all levels and renovation of 35,000 square feet of research space. Research currently encompasses six main themes: Development, Cell Biology, Genomics, Neuroscience, Metabolism, and Proteomics/Glycomics.
Applicants should upload the following items to https://academicjobsonline.org/ajo/jobs/25282 :
- Cover letter (max 1 page),
- Curriculum vitae including full list of publications,
- Research statement with a description of significant research accomplishments and future research plans (max 3 pages)
- Diversity, Equity, and Inclusion (DEI) statement highlighting the commitment to mentoring diverse trainees and contributing to increasing inclusiveness and diversity in the Department (max 1 page)
- Names and email addresses of three persons who will provide letters of recommendation (to be submitted online by the reference writers)
Applications will be reviewed on a rolling basis through the Fall until the position is filled. Please direct email queries to biocb@bu.edu.
Boston University is an equal opportunity and affirmative action employer
New Publication from the Garcia-Marcos Lab: Coordinating signaling responses in neurons
The Garcia-Marcos Lab has recently published a study in Molecular Cell (https://doi.org/10.1016/j.molcel.2023.06.006; prepint in: https://www.biorxiv.org/content/10.1101/2023.03.03.529094v1) titled “Fine-tuning GPCR-mediated neuromodulation by biasing signaling through different G protein subunits”. The paper describes how different signaling responses triggered by the same neurotransmitter receptor must be carefully scaled to ensure proper brain function. They found that the protein named GINIP shifts the balance of two different G protein sub-species activated simultaneously by G protein-coupled receptors (GPCRs), a large family of surface receptors that respond many neurotransmitters and neuropeptides, including GABA, dopamine, serotonin, or opioids. This mechanism operates in synapses that dampen neurotransmission and, when disabled, results in increased seizure susceptibility in mouse models. These findings have important implications for the fundamental understanding of neuronal communication and for the development of new therapeutic agents that act on GPCRs.
This work was co-led by Jong-Chan Park (Postdoc) and Alex Luebbers (Graduate Student) with collaborations from the Martemyanov Lab at UF Scripps Biomedical Research Institute and the Yano Lab at Northeastern University, and has been highlighted by Molecular Cell (https://doi.org/10.1016/j.molcel.2023.06.034) and Science Signaling (https://www.science.org/doi/10.1126/scisignal.adj6131).
Grishok Lab recently (June 24-28, 2023) attended the 24th International C. elegans Conference at the Scottish Event Campus (SEC) in Glasgow.
Grishok Lab recently (June 24-28, 2023) attended the 24th International C. elegans Conference at the Scottish Event Campus (SEC) in Glasgow.

Gian Sepulveda and Thomas Liontis were busy presenting posters.
Glasgow's medieval history is impressive, and so is modern mural art. 
