Mikel Garcia-Marcos Promotion
The Department would like to congratulate Dr. Mikel Garcia-Marcos on his recent promotion to Associate Professor of Biochemistry. This is a well-deserved honor!
Inhibiting G protein signaling and disease
Citation: Specific inhibition of GPCR-independent G protein signaling by a rationally engineered protein.
Leyme A, Marivin A, Maziarz M, DiGiacomo V, Papakonstantinou MP, Patel PP, Blanco-Canosa JB, Walawalkar IA, Rodriguez-Davila G, Dominguez I, Garcia-Marcos M. Proc Natl Acad Sci U S A. 2017 Nov 13. pii: 201707992. doi: 10.1073/pnas.1707992114. [Epub ahead of print] PMID: 29133411
Center for Network Systems Biology Grand Opening
Andrew Emili Heads New Center for Network Systems Biology
from BU Today
10.03.2017  Andrew Emili, with a joint appointment as a professor in the MED biochemistry department and the CAS biology department, is the director of the new University-wide Center for Network Systems Biology. Photo by Cydney Scott
Andrew Emili, with a joint appointment as a professor in the MED biochemistry department and the CAS biology department, is the director of the new University-wide Center for Network Systems Biology. Photo by Cydney Scott
As a McGill University undergraduate, Andrew Emili earned money putting together IKEA furniture. The assembly instructions may have stymied his customers, but at least instructions existed. That’s more than can be said for Emili’s current challenge: mapping the network of interactions between the tens of thousands of proteins encoded in the human genome.
“It’s like trying to put together IKEA furniture when you’ve lost the assembly instructions,” says Emili, a molecular systems biologist who arrived at Boston University from the University of Toronto in July. “You see bolts, you see holes, and you know the relationship between the two, but not which ones go where.”
Emili, jointly appointed to the School of Medicine biochemistry department and the College of Arts & Sciences biology department, is director of the new University-wide Center for Network Systems Biology (CNSB).
Human health and development depend on the network of protein interactions, he says. Yet despite rapid advances in genomics, scientists know little about how these interactions work and how faulty interactions lead to disease. He uses proteomics, the study of the protein products of genes, and mass spectrometry, a tool that can separate individual proteins from their connections, as well as bioinformatics and other molecular genetic and genomic technologies, to create maps of protein interactions. He then makes his maps, which he describes as assembly instructions for molecular networks, available to the broader research community. His ultimate goal, he says, is to translate this basic knowledge into novel diagnostic and therapeutic tools for cancer, cardiovascular disease, and Alzheimer’s disease and other neurodegenerative disorders.
Emili’s vision for the CNSB, he says, is “to create a highly collaborative, multidisciplinary research hub that tackles important fundamental questions in the field by forging new links with interested researchers across both BU campuses, the greater Boston area, and the world.”
The departments of biochemistry and biology will host a reception today on the Medical Campus—open to students and faculty from both campuses—to mark the opening of the center and Emili’s appointment as director, at the Silvio O. Conte Medical Research Center, 71 E. Concord St., from 3 to 5 p.m.
Emili is widely regarded as a leader in proteomics, mass spectrometry, and network systems biology, says David Harris, a MED professor and chair of biochemistry. He says Emili’s work with mass spectrometry will complement that of Catherine E. Costello, a William Fairfield Warren Distinguished Professor, a MED professor of biochemistry, and director of the Center for Biomedical Mass Spectrometry. With Emili’s arrival, Harris says, “we have an incredibly strong presence in multiple kinds of mass spectrometry.”
While the CNSB, as well as Emili’s lab, will be housed within the biochemistry department, he will serve as a bridge between the Medical and Charles River Campuses and will also have an office in the Life Sciences and Engineering Building—and eventually some lab space—at 24 Cummington Mall and teach classes in the biology department.
“He will collaborate widely across the University,” says Harris. “This recruitment from the start was a joint effort of the medical school and the Charles River Campus.”
“It’s really great to have him come in and be a leader,” says Kim McCall, a CAS professor and chair of biology, noting that her department has recently hired three junior faculty in systems biology. “His research is highly collaborative. He’s already talking to people in chemistry. He’s done work related to evolution, so that bridges with our scientists who are doing evolutionary biology. He’ll be important to bioinformatics on the CRC as well.”
A map of human protein interactions, from a 2012 Cell study, a collaboration between Emili and Edward Marcotte of the University of Texas, Austin. The spheres, or dots, are proteins; the lines are interactions between proteins. Emili explains: “The network layout reflects local clustering of proteins to form specific macromolecules—stable complexes—while the broader connectivity between these assemblies shows crosstalk underlying biological circuits and cellular processes. Many of the complexes identified in the study were previously unknown and/or have links to human disease, providing valuable insights into pathobiological mechanisms.” Image courtesy of Emili
“A Google or Facebook of biology”
“If you really want to understand the cell,” Emili says, “you have to understand what the protein molecules do—how they interact, what their functions are, how they’re regulated. The DNA is the code or the raw information, but the proteins are the molecules that are the building blocks. They’re not just abstract information. You can think of them as the construction workers in the cell.”
He thinks of his team of researchers, he says, “as a Google or Facebook of biology,” mapping social networks of proteins that provide clues to how proteins function. “Proteins interact functionally and physically in very dynamic and intriguing ways,” he says. “It’s about who knows who and who’s connected to whom.
“In a disease state, these networks are often perturbed or modified or they fail in some way,” he says, “and if we want to reverse a disease or prevent it, we have to understand how the networks go awry and what something looks like when it’s broken and what it looks like when it’s not broken.”
In 2015, Emili and Edward Marcotte, a University of Texas, Austin, professor of molecular biosciences, led a landmark Nature study that revealed tens of thousands of new protein interactions across nine animal species—baker’s yeast, amoebas, sea anemones, flies, worms, sea urchins, frogs, mice, and humans. Using mass spectrometry to analyze cell samples from each species, the researchers found which proteins worked together in networks and compared their structures across species. Their map provided clues to how these protein associations evolved over time.
“Andrew’s work is highly relevant to a broad range of questions in both basic and applied biomedical research,” says Michael Sorenson, a CAS professor of biology. As an evolutionary biologist, Sorenson says, he particularly appreciates Emili’s 2015 Naturestudy and how it “beautifully illustrates the way in which animal diversity has evolved by building upon and tweaking a common set of fundamental cellular processes that has functioned in much the same way for a billion years or more.”
Emili came to BU from the University of Toronto, where he was a professor of molecular genetics and the Council of Ontario Universities Ontario Research Chair in Biomarker Discovery. He was also a principal investigator and a founding member of the Donnelly Centre for Cellular and Biomolecular Research. He earned a PhD in molecular and medical genetics from the University of Toronto in 1997 and pursued postdoctoral studies as a Damon Runyon/Walter Winchell Cancer Research Fellow with cell geneticist Leland Hartwell, a Nobel laureate, at the Fred Hutchinson Cancer Research Center in Seattle, where Hartwell is president and director emeritus. During that same period, Emili learned protein mass spectrometry with John Yates III, then a University of Washington School of Medicine associate professor of molecular biotechnology, now a Scripps Research Institute professor of chemical physiology.
“BU has tremendous resources,” says Emili, “and given the widespread community support, I think the center can leapfrog ahead and chart out some exciting new terrain to claim and explore.
“Let’s see what riches this inititiative will yield.”
The School of Medicine biochemistry department and College of Arts & Sciences biology department will host a reception, open to students and faculty from both the Charles River Campus and the Medical Campus, to mark the opening of the Center for Network Systems Biology and Andrew Emili’s appointment as director, today, Tuesday, October 3, from 3 to 5 p.m., at the Silvio O. Conte Medical Research Center, 71 E. Concord St., on the Medical Campus.
GIV is Druggable
Earlier this year, the Garcia-Marcos laboratory reported detailed structural information describing how trimeric G proteins are activated by GBA motifs, protein segments capable of triggering G-protein signaling by a GPCR-independent mechanism. The work focused on GIV, a nucleotide exchange factor for Gαi3. Because we had previously shown that the GIV-Gαi3 interaction is required for cancer metastasis, we investigated if it could be disrupted by small molecules. The identification of such compounds would represent the first step in the development of novel anti-metastatic drugs, an urgently needed arm of current cancer therapeutic strategies.
Disrupting protein-protein interactions (PPIs) like the one established by GIV and Gαi3, however, is notoriously challenging. A significant hurdle to therapeutic development is demonstrating that a given PPI can be targeted by small molecules in the first place – i.e. they tend not to be “druggable.” To establish the druggability of our target, we combined computational approaches and wet laboratory techniques, drawing on insights gathered from our recent studies. We concluded disruption of the PPI target could indeed be achieved by small molecules and furthermore that the mode of action can be readily predicted by utilizing structural information.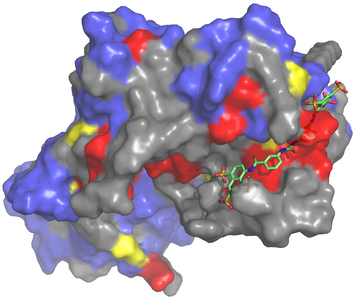
The work establishes a robust pipeline for the discovery and validation of inhibitors of the GIV-Gαi3 interface and identifies a small molecule that can serve in such a role. A limitation is that the small molecule we validated was not suitable for experimentation in cancer cells or patients. The study nonetheless provides an important proof of principle for the druggability of our target, success with which could be achieved by screening larger libraries of chemical compounds. Such high-throughput screens are currently underway in our laboratory.
This work involved collaboration with the group of Francisco J. Blanco, from the CIC-BioGUNE in Spain and was published in the journal Scientific Reports.
Reference
The Gαi-GIV binding interface is a druggable protein-protein interaction. DiGiacomo V, de Opakua AI, Papakonstantinou MP, Nguyen LT, Merino N, Blanco-Canosa JB, Blanco FJ, Garcia-Marcos M. Sci Rep. 2017 Aug 17;7(1):8575. doi: 10.1038/s41598-017-08829-7. PMID: 28819150
Faculty Appointed to NIH Study Sections
Dr. Konstanin Kandror was appointed to the National Institutes of Health, Cellular Aspects of Diabetes and Obesity (CADO) study section and Dr. Mikel Garcia-Marcos was appointed to the Molecular and Integrative Signal Transduction study section. These appointments are made based on "their demonstrated competence and achievement in their scientific discipline as evidenced by the quality of research accomplishments, publications in scientific journals, and other significant scientific activities, achievements and honors."
New Research Discoveries: Prion proteins and neuron degeneration
A new paper in eLife from the Harris lab has uncovered a novel function for different domains of the prion protein. Bei Wu, an Instructor in the laboratory was lead author.
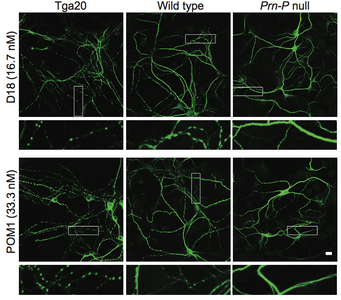
Prion diseases, or transmissible spongiform encephalopathies, comprise a group of fatal neurodegenerative disorders in humans and animals for which there are no effective treatments or cures. These diseases are caused by refolding of the cellular prion protein (PrPC) into an infectious isoform (PrPSc) that catalytically templates its abnormal conformation onto additional molecules of PrPC.A similar, prion-like process may play a role in other neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases and tauopathies, which are due to protein misfolding and aggregation. Here, using a combination of electrophysiological, cellular, and biophysical techniques, we show that the flexible, N-terminal domain of PrPC functions as a powerful toxicity-transducing effector whose activity is tightly regulated in cis by the globular C-terminal domain. Ligands binding to the N-terminal domain abolish the spontaneous ionic currents associated with neurotoxic mutants of PrP, and the isolated N-terminal domain induces currents when expressed in the absence of the C-terminal domain. Anti-PrP antibodies targeting epitopes in the C-terminal domain induce currents, and cause degeneration of dendrites on murine hippocampal neurons, effects that entirely dependent on the effector function of the N-terminus. NMR experiments demonstrate intramolecular docking between N- and C-terminal domains of PrPC, revealing a novel auto-inhibitory mechanism that regulates the functional activity of PrPC.
Welcome New Faculty
We are delighted to announce the recruitment of 2 new faculty members who will be joining the Department on July 1, 2017. The search that identified these faculty members was a joint effort between the Biochemistry Department and the Genome Science Institute.
Nelson Lau, Ph.D., will be appointed as an Associate Professor. His laboratory will be located on the second floor of the K Bldg. Nelson has a long-standing involvement in the RNA field. He completed his Ph.D. at MIT in 2004 with Dr. David Bartel, a pioneer in the then nascent field of microRNAs. As part of his thesis work, he cloned the first large collection of microRNAs from C. elegans, work that was awarded the Newcomb Cleveland prize from the AAAS in 2002. From 2004-2009, Nelson was a Helen Hay Whitney Foundation fellow in the laboratory of Dr. Robert Kingston at Massachusetts General Hospital/Harvard Medical School. During his fellowship, he was the first to describe the piRNA complex from rats and mice, a discovery that was a runner-up for Science magazine’s 2006 Breakthrough of the Year. In 2009, Nelson was recruited as an Assistant Professor to the Department of Biology at Brandeis University. There, he established a vibrant and productive research program focused on regulation of the genome by transposon landscapes and the Piwi/piRNA pathway. His laboratory produced a number of important advances, including: (1) Establishment of new links between the Piwi pathway, transposon landscapes, and long non-coding RNAs; (2) Discovery of eutherian-mammal conserved genic piRNA clusters; (3) Development of new technical methods to study the RNAi pathway. Nelson’s interests complement those of Alla Grishok and Daniel Cifuentes, further adding to our department’s strengths in RNA biology. Nelson also adds two new model organisms to our department: Drosophila and Xenopus tropicalis.
Andrew Emili, Ph.D., will be a Professor, with dual appointments in the Dept. of Biochemistry and in the Department of Biology on the Charles River Campus. He will establish a Center for Network Systems Biology, which will be located on the third floor of the K Bldg. in space that is currently being renovated for this purpose. Andrew was recruited through the Provost’s Senior Faculty Hiring Initiative, aimed at attracting world-class researchers to Boston University who will bridge the two campuses. He is currently a Professor in the Donnelly Centre for Cellular and Biomolecular Research and the Department of Molecular Genetics at the University of Toronto, where he has been located since 2000. He received his Ph.D. (1997) in Molecular and Medical Genetics from the University of Toronto, and was a postdoctoral fellow (1997-2000) at the Fred Hutchison Cancer Research Center in Seattle working with John Yates. Andrew is an international leader in the analysis of protein interaction networks. He uses systems-level analysis, bioinformatics and especially proteomics to answer large-scale questions about protein-protein interaction networks in cells. Andrew’s publication list includes high profile, proteome-wide studies of protein complexes in yeast, E. coli, and human cells, and his group has documented hundreds of novel complexes linked to development and disease. Andrew’s center will synergize with the Center for Biomedical Mass Spectrometry, directed by Cathy Costello and Joe Zaia, further establishing our dept. as a leader in applications of mass spectrometry to biological problems.
Dahoud Breast Cancer Pilot Awards
Congratulations to several Biochemistry faculty who were recently awarded Dahoud Breast Cancer Pilot Awards.
Alla Grishok, PhD, Associate Professor of Biochemistry, Dafne Cardamone, PhD, Instructor, and Catherine Costello, PhD, Director of Center for Biomedical Mass Spectrometry, will study regulation of cancer-promoting Myc protein using a model metastatic breast cancer cell line. Myc binds DNA and activates a large network of genes that together transform normal cells into cancer cells. Myc activity is elevated in most human cancers and is especially relevant for Myc-driven triple (estrogen, progesterone and Her2) negative breast cancer. Dr. Grishok and colleagues will investigate new mechanisms that increase Myc protein activity: 1) adding specific sugar residues, and 2) protein truncation. New compounds that directly inhibit Myc or inhibit enzymes that activate Myc could be developed into new cancer therapies.

Xaralabos Varelas, PhD, Associate Professor of Biochemistry and Stefano Monti, PhD, Associate Professor of Medicine and Biostatistics, will study the causes of aggressive triple negative breast cancers. The team will determine how abnormal signaling networks drive gene expression changes that lead to aggressive breast cancers and then categorize subsets of aggressive breast cancers, thereby better targeting the most effective treatments based on the genes expressed in the tumor.

Mikel Garcia-Marcos Awarded Grunebaum Fellowship
Mikel Garcia-Marcos has been selected as the Karin Grunebaum Cancer Research Fellow for a second year in a row. The Grunebaum Faculty Research Fellowship is a BUSM annual faculty award that provides $25,000 in total funds to a selected faculty member for a period of one year. Several Faculty members of our Department, including Bob Varelas and Valentina Perissi, have been awarded this fellowship in the past. Dr. Garcia-Marcos plans to work on developing a novel therapeutic strategy against cancer metastasis that targets an unconventional mechanism of heterotrimeric G protein activation not mediated by surface receptors (GPCRs).
Cathy Costello receives prestigious mass spectrometry award
Cathy Costello was recently awarded the 2017 Award for a Distinguished Contribution in Mass Spectrometry by the American Society for Mass Spectrometry. This award recognizes a singular significant achievement and was for her pioneering contributions to the development of tandem mass spectrometry of glycans and glycoconjugates. Addtional details of the award can be found on the ASMS web page. Congratulations Cathy!
