
Xiaohu Mei, Ph.D.
Instructor of Pharmacology, Physiology & Biophysics
Research
Cardiovascular disease remains the leading cause of death in United States. Apolipoprotein A-I (apoA-I) is the major protein in high-density lipoprotein (HDL) and plays a vital role during the process of reverse cholesterol transport (RCT). Knowledge of the high-resolution structure of full-length apoA-I is essential for a molecular understanding of the function of HDL at various steps during the RCT pathway. Due to the flexible nature of apoA-I and inherent aggregation properties, the structure of full-length apoA-I has evaded description for over three decades. We use the techniques of modern molecular biophysics and structural biology to study the structure and function of apoA-I. We primarily rely on protein crystallography and molecular modeling coupled with SAXS to study the structure of apoA-I. In addition, circular dichroism, fluorescence microscope, electron microscopy, NMR and cellular assay are used to understand the function of apoA-I.
I. Crystal structure of N-terminal domain of apoA-I
We have successfully solved the crystal structure of C-terminal truncated apoA-I (Δ(185-243)) at 2.2Å resolution (PDB: 3R2P). The structure shows that it forms a half circle of dimer. The N-terminal domain of each molecule forms a four-helix bundle with the helical C-terminal region of the symmetry mate. The structure suggests the role of dimerization during the HDL formation and allows us to model the formation of discoid HDL particles in three steps.
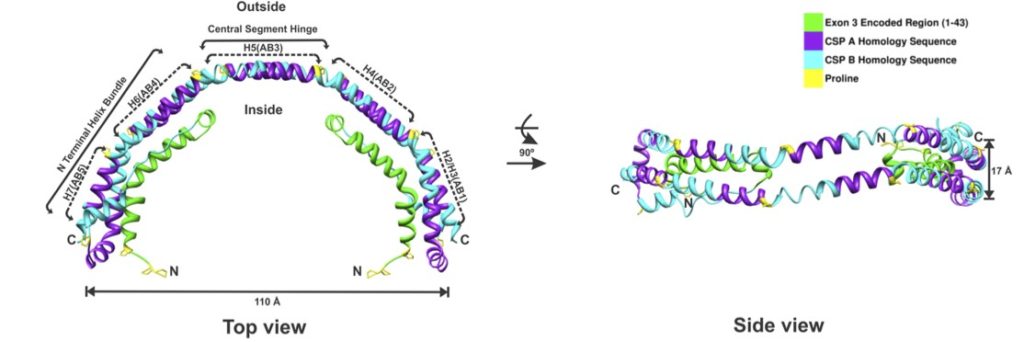
II. Probing the C-terminal domain of apoA-I
To understand the role of the C-terminal domain, constructs with sequential elongation of Δ(185-243), by increments of 11-residue sequence repeats were studied and compared with Δ(185-243) and WT apoA-I. Constructs up to residue 230 showed progressively decreased percent α-helix with similar numbers of helical residues, similar detergent and lipid binding affinity, and exposed hydrophobic surface. These observations suggest that the C-terminal domain is unstructured with the exception of the last 11-residue repeat (H10B). Similar monomer-dimer equilibrium suggests that the H10B region is responsible for nonspecific aggregation. Cholesterol efflux progressively increased with elongation in the absence of the H10B. In summary, the sequential repeats in the C-terminal domain are probably unstructured with the exception of H10B. This segment appears to be responsible for initiation of lipid binding and aggregation, as well as cholesterol efflux, and thus plays a vital role during HDL formation. Based on these observations and the Δ(185-243) crystal structure, we propose a lipid-free apoA-I structural model in solution and update the mechanism of HDL biogenesis.

III. Conversion between monomer/dimer states
Based on the crystal structure, the H5 (residues 121-142) region is the most flexible region in the structure. Indeed, residues 136–141 exist in a poorly folded “helix-like” conformation. These observations lead us to suggest that the H5 may function as a hinge in the monomer form of Δ(185–243)apoA-1 in dilute solution. The C-terminus folds back to replace the corresponding segments from the opposing molecule of the dimer. Our crystal structure together with experiments from other labs substantiates the two-domain structure of apoA-I in solution with H5 functioning as a hinge determining the monomer to dimer conversion with protein concentration. Mutation studies are underway to test the hypothesis.
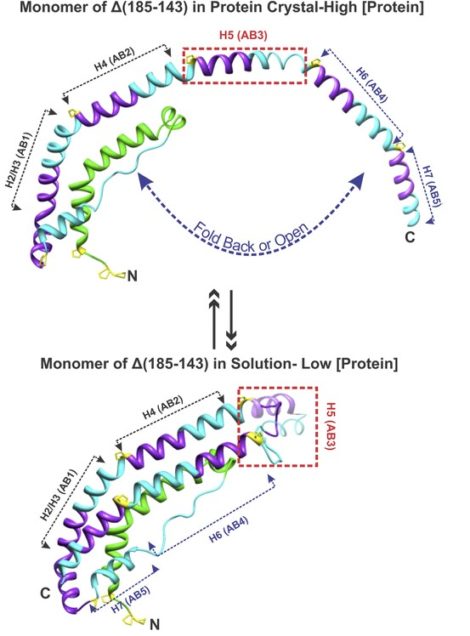
Our long-term goal is to achieve the high-resolution structure determination of full-length apoA-I in monomer and dimer states thus understand the conversion mechanism between different states, which will help to reveal the HDL formation mechanism and function during the RCT process. This may lead to more effective prevention and treatment of cardiovascular disease.
Selected Publications
Mei X., Liu M., Herscobitz H., Atkinson D.(2016) Probing the C-terminal domain of lipid-free apoA-I demonstrates the vital role of the H10B sequence repeat in HDL formation. J Lipids Res 57(8):1507-17. (PMID: 27317763)
Mei X., Atkinson D.(2015) Lipid-free Apolipoprotein A-I Structure: Insights into HDL Formation and Atherosclerosis Development. Arch Med Res 46(5):351-60. (PMID: 26048453)
Mei X., Atkinson D.(2011), Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high-density lipoprotein (HDL) by dimerization. J Biol Chem 286(44):38570-82. (PMID: 21914797)
Links:
Faculty Profile
ResearchGate
PubMed search for Dr. Mei
Contact Us
Department of Pharmacology, Physiology & Biophysics
Chobanian & Avedisian School of Medicine
700 Albany Street, W330A
Boston MA 02118-2526
Phone:(617)358-8491
e-mail: meixiao@bu.edu