Two new publications from the Garcia-Marcos Lab
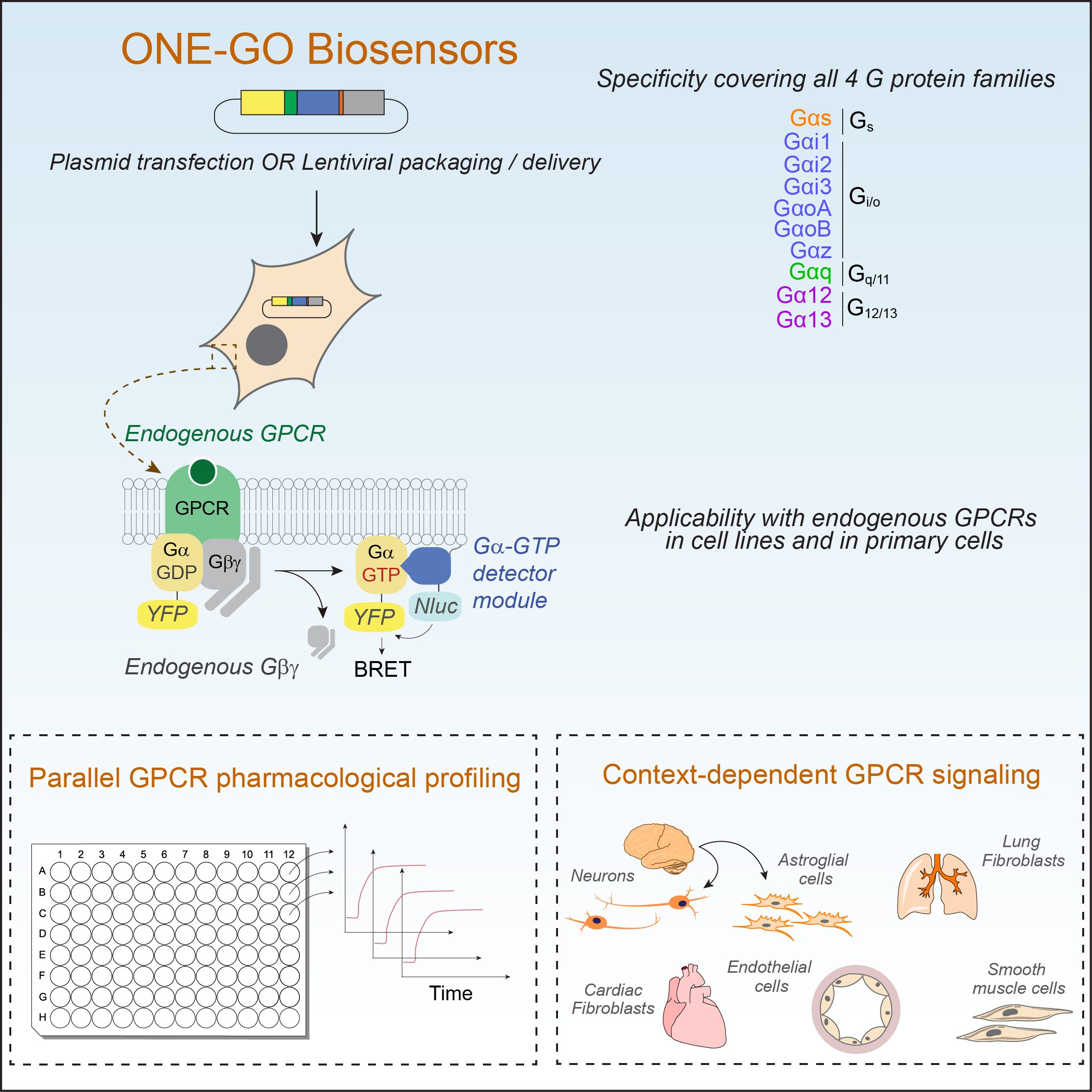
In a first publication, the Garcia-Marcos Lab has recently published the development and implementation of a suite of G protein biosensors broadly applicable to detect GPCR activity in scalable assay formats and in physiologically relevant systems like primary cells. By directly measuring endogenous GPCR activity, these biosensors, named ONE-GO, reveal that responses are frequently dependent on the cell context or state. This resource article has been published in the journal Cell (https://www.sciencedirect.com/science/article/pii/S0092867424000655?via%3Dihub) and all related plasmid reagents are deposited in Addgene (https://www.addgene.org/kits/garcia-marcos-one-go-biosensors/). The work in this paper was led by current PhD student Remi Jnaicot and former postdoc Marcin Maziarz, and includes collaborations with the Layne Lab in our Department and the Wu lab at Stanford.
In a second paper, Mikel has reviewed a lot of the research related to his lab's work on atypical mechanisms of signaling over the last 15 years.G proteins are molecular switches that relay signals from G protein-coupled receptors across the cell membrane. While it is generally assumed that G proteins, molecular switches that trasnduce signals, are activated exclusively by G protein-coupled receptors (GPCRs), this review summarizes the mounting evidence showing that G proteins can signal via a class of G protein regulator proteins that contain a Gα-binding-and-activating (GBA) motif. The mechanisms and structural basis for this GPCR-independent mechanism of G protein signaling are presented, as well as a description of strategies to manipulate GBA-G protein coupling for research purposes or the potential development of therapeutics. This paper has been published as a commissioned review in the Journal of Biological Chemistry (https://www.sciencedirect.com/science/article/pii/S0021925824001327?via%3Dihub).
Mikel Garcia-Marcos elected Chair of Division of National Scientific Society

Mikel Garcia-Marcos has been elected as Chair of the Division of Molecular Pharmacology of the American Cancer Society of Pharmacology and Experimental Therapeutics (ASPET).
He will serve a 1-year term as Chair-Elect followed by a 1-year term as Chair. In this role, he will oversee the activities of the Executive Committee involving planning of the Society's Annual Meeting, financially supporting external scientific meetings, selection of scientific achievement awardees, and organizing other activities like online webinars and workshops.
The American Society for Pharmacology and Experimental Therapeutics (ASPET) is a 4,000 member scientific society whose members conduct basic and clinical pharmacological research and work for academia, government, large pharmaceutical companies, small biotech companies, and non-profit organizations. ASPET is a global pharmacology community that advances the science of drugs and therapeutics to accelerate the discovery of cures for disease.
New publication sheds light onto the molecular basis of neurotransmitter signaling
A new paper led by Alex Luebbers in the Garcia-Marcos Lab dissects the molecular basis for how GINIP, a Gαi-interacting protein in neurons involved in controlling pain and seizures, modulates GPCR responses triggered by neurotransmitters. GINIP mimics how G-protein signaling effectors bind to Gα subunits via its PHD domain to differentially scale discrete G-protein signaling branches. The paper has been published in Structure (https://www.sciencedirect.com/science/article/abs/pii/S0969212623003891?via%3Dihub), and was previously posted on bioRxiv (https://www.biorxiv.org/content/10.1101/2023.04.20.537566v1).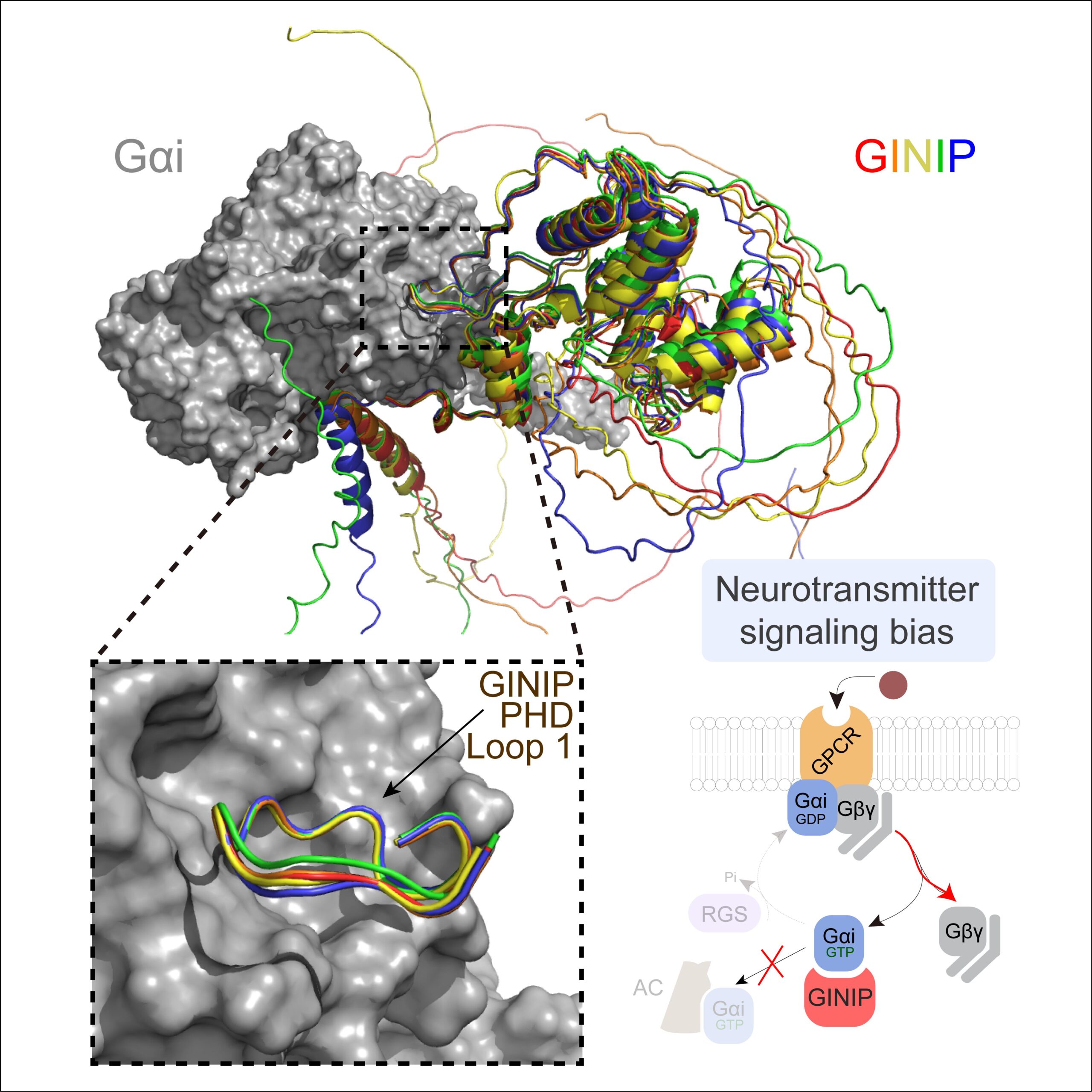
This paper builds and explands on another paper recently published in the same lab establishing that the neuronal protein GINIP shapes GPCR inhibitory neuromodulation via a unique mechanism of G-protein regulation that controls pain and seizure susceptibility (Park, Luebbers, et al Molecular Cell, 2023). However, the molecular basis of this mechanism remained ill-defined because the structural determinants of GINIP responsible for binding and regulating G proteins were not known. The newly published paper combined hydrogen-deuterium exchange mass spectrometry, computational structure predictions, biochemistry, and cell-based biophysical assays to demonstrate an effector-like binding mode of GINIP to Gαi. These findings explain the molecular basis for a post-receptor mechanism of G-protein regulation that fine-tunes inhibitory neuromodulation.
This paper features collaborations with the labs of Stephen Eyles at UMass-Amherst and of Joshua Levitz at Weill Cornell.
New Publication from the Garcia-Marcos Lab: Coordinating signaling responses in neurons
The Garcia-Marcos Lab has recently published a study in Molecular Cell (https://doi.org/10.1016/j.molcel.2023.06.006; prepint in: https://www.biorxiv.org/content/10.1101/2023.03.03.529094v1) titled “Fine-tuning GPCR-mediated neuromodulation by biasing signaling through different G protein subunits”. The paper describes how different signaling responses triggered by the same neurotransmitter receptor must be carefully scaled to ensure proper brain function. They found that the protein named GINIP shifts the balance of two different G protein sub-species activated simultaneously by G protein-coupled receptors (GPCRs), a large family of surface receptors that respond many neurotransmitters and neuropeptides, including GABA, dopamine, serotonin, or opioids. This mechanism operates in synapses that dampen neurotransmission and, when disabled, results in increased seizure susceptibility in mouse models. These findings have important implications for the fundamental understanding of neuronal communication and for the development of new therapeutic agents that act on GPCRs.
This work was co-led by Jong-Chan Park (Postdoc) and Alex Luebbers (Graduate Student) with collaborations from the Martemyanov Lab at UF Scripps Biomedical Research Institute and the Yano Lab at Northeastern University, and has been highlighted by Molecular Cell (https://doi.org/10.1016/j.molcel.2023.06.034) and Science Signaling (https://www.science.org/doi/10.1126/scisignal.adj6131).
New Publication: The Garcia-Marcos lab, in collaboration with members of the Department of Chemistry at Boston University and the CSIC-CIB in Spain, have published in Proceeding of the National Academy of Sciences (PNAS) …
The Garcia-Marcos lab, in collaboration with members of the Department of Chemistry at Boston University and the CSIC-CIB in Spain, have published in Proceeding of the National Academy of Sciences (PNAS) the discovery of a chemical compound that specifically blocks an aberrant
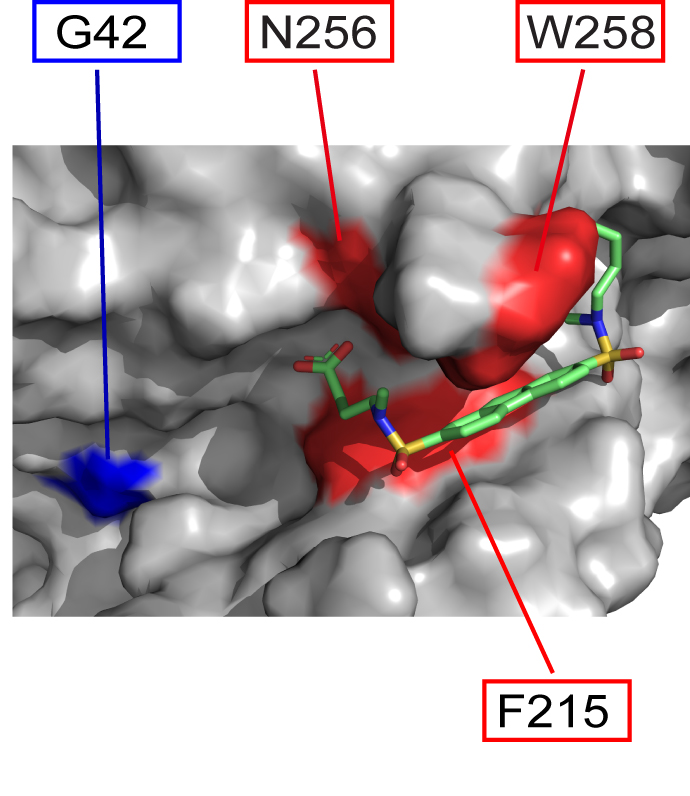 mechanism of signaling in cancer cells that drives invasion and metastasis. The publication can be found here: https://www.pnas.org/doi/10.1073/pnas.2213140120
mechanism of signaling in cancer cells that drives invasion and metastasis. The publication can be found here: https://www.pnas.org/doi/10.1073/pnas.2213140120
Human diseases frequently arise from defects in the mechanisms by which external cues are sensed and relayed to the interior of the cell. The proteins most widely targeted by existing therapeutic agents belong to a large family of cell surface receptors named G-protein-coupled receptors (GPCRs), which relay external cues by activating G-proteins in the interior of cells. Here, we report the surprising discovery of a synthetic small molecule that selectively targets G-proteins without compromising their ability to relay signals from GPCRs. Instead, this small molecule disrupts an atypical, GPCR-independent mechanism of G-protein signaling involved in cancer. This work reveals an alternative paradigm in targeting components of a signaling machinery with broad relevance in cellular communication in health and disease.
Research discoveries: Daple orchestrates actomyosin assembly
Congratulations to Arthur Marivin (Garcia-Marcos lab) for his new paper in the Journal of Cell Biology describing how association of DAPLE with PAR polarity complexes at cell-cell junctions coordinates actomyosin assembly in epithelial cells.
Jingyi Zhao, Ph.D (Garcia-Marcos Lab) awarded 2022 Dahod International Scholar
Congratulations to Jingyi Zhao, Ph.D. (Garcia-Marcos Lab) who was awarded 2022 Dahod International Scholar.
Jingyi, a postdoc in Dr. Mikel Garcia-Marcos’ laboratory, will characterize a small molecule inhibitor of a novel signaling mechanism that promotes breast cancer metastasis. He has identified a promising candidate molecule, named IGGi-11, that disrupts a protein signaling complex specifically present in metastatic cancer cells. He intends to further confirm the specificity of this molecule in preventing cancer cell invasiveness without overt cytotoxic effects in normal cells, and to work towards developing analog compounds with improved properties in preclinical breast cancer models.
Remi Janicot awarded American Heart Association fellowship
Remi Janicot in the Garcia-Marcos laboratory was awarded an American Heart Association (AHA) predoctoral fellowship for his project "Optical biosensor platforms for the direct interrogation of GPCR signaling in cardiovascular cells”
The goal of this project is to create new experimental tools that can transform how GPCRs are studied. The main benefit over current methods is that these tools can be easily used in cell models that are relevant to study heart or lung disease, which has been an important limitation in the field. These new experimental possibilities would open new doors for the entire cardiovascular research community. The tools designed in this project will allow to study GPCRs with fidelity and precision. This would pave the way to develop new drugs to treat life-threatening cardiac disorders.
Congratulations Remi!
Mikel Garcia-Marcos receives John Abel Award from ASPET
Congratulations to Dr. Mikel Garcia-Marcos for receiving the 2022 American Society for Pharmacology and Experimental Therapeutics (ASPET) John J. Abel Award in Pharmacology. The Abel Award is named after the founder of ASPET. It was established in 1946 to stimulate fundamental research in pharmacology and experimental therapeutics by young investigators.
The award will be presented at the ASPET Business Meeting and Awards Presentation during the ASPET Annual Meeting at Experimental Biology 2022 on Saturday, April 2 at 4:30 pm in Philadelphia. Additionally, Dr. Garcia-Marcos will deliver the Abel Award Lecture titled The Secret Life of G Proteins to open the 2022 annual meeting on Saturday, April 2 at 10:00 am in Philadelphia.
Congratulations to Mikel Garcia-Marcos on his promotion to Full Professor of Biochemistry!
Congratulations to Mikel Garcia-Marcos on his promotion to Full Professor of Biochemistry!

The Garcia-Marcos Lab investigates signal transduction mechanisms with the ultimate goal of elucidating the molecular basis of human diseases and developing novel therapeutic approaches.