BUPID-XL
Chemical cross-linking combined with mass spectrometry (XL-MS) is a powerful technique for analyzing protein-protein interactions and gathering structural information. Interpretation of the results of these experiments can be challenging due to the larger search spaces compared with those in conventional proteomics experiments. Several software packages have become available in recent years but many have limitations on what data they can process corresponding to certain classes of XL-MS experiments such as ones involving isotopic labels. We developed BUPID-XL which can interpret data from a variety of different cross-linkers and thus facilitates the penetration of XL-MS techniques into a greater number of mass spectrometry laboratories.
The BUPID-XL software package consists of several components, each one responsible for identifying peptides linked with a different cross-linker. These typically fall into two categories: cross-linkers that are cleavable and non-cleavable by MS, with some variations depending on other factors specific to their chemistries. By submitting raw spectra as mzML along with some analysis parameters, the data will be analyzed using the appropriate tools and the results will be returned as a CSV formatted file. The results file includes information about the linked peptides, modifications, and scoring metrics.
Architecture
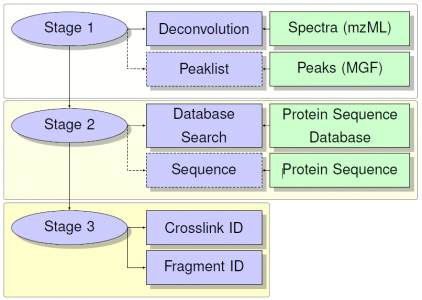
Peptides are identified initially with a precursor mass comparison. These IDs are filtered based on the MS/MS. MS/MS scans are first deconvoluted using the deconvolution tool developed in-house for BUPID Top-Down framework. Filtering is then accomplished using one of several available methods depending on the properties of the cross-linker. Processing jobs for data containing cleavable linkers focus on identifying the intact peptides with modifications consisting of the linker fragments and any user provided modifications. Tools for different classes of linkers then proceed to identify peptide backbone fragmentation, with or without the linker present, to serve as confirmation for the precursor mass ID.
BUPID-XL has been tested using HCD spectra of individual proteins and protein complexes treated with different cross-linking reagents and represent relevant ongoing research projects in our center. The software was able to identify peptides from these proteins linked with several different cleavable and non-cleavable cross-linkers.
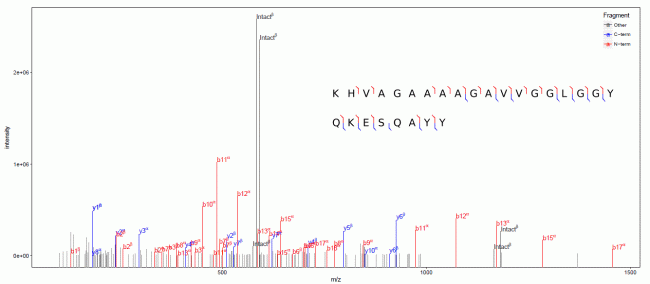
Example output from BUPID-XL showing cross-linked peptides.