News
August 2018
Colon cancer is the second leading cause of cancer deaths in non-smoking men and women, and because surgery has limited therapeutic role with metastatic colon cancer, chemotherapy is widely used as the first-line therapy. However, several studies have indicated that patients harboring SMAD4 mutations corresponding to chromosome 18q deletions that frequently occur during advanced stages of colon cancer exhibit resistance to chemotherapy. Dr. Thiagalingam has been awarded a BU-CTSI Integrated Pilot grant entitled “Sensitization of chemotherapy resistant metastatic colon cancer” to initiate the identification of molecular mediators that promote colon cancer metastasis and resistance to common chemotherapeutic agents correlating to the loss of function of SMAD4 for precision targeting to restore sensitivity to chemotherapy. In the long-term, this award will help Dr. Thiagalingam to generate critical preliminary data to tap into funds to undertake comprehensive understanding of resistance to chemotherapy in colon cancer.
July 2018
Dr. Huiping Zhang receives BU CTSI grant to study the epigenetic mechanism of cocaine use disorders (CUDs)
Dr. Huiping Zhang receives BU CTSI grant to study the epigenetic mechanism of cocaine use disorders (CUDs). He and his collaborator Dr. Vidhya Kumaresan will investigate cocaine use/withdrawal induced microRNA (one type of epigenetic markers) expression alterations in the brain’s reward circuit (e.g., the nucleus accumbens) using a translational rat model of incubated drug craving (i.e., extended access to cocaine with prolonged abstinence and consequent increased craving). It is expected that cocaine withdrawal responsive microRNAs (or their target genes) could be potential biomarkers for CUDs and novel therapeutic targets.”
May 2018
Shoumita Dasgupta Elected to Association of Professors of Human and Medical Genetics
Shoumita Dasgupta, PhD, associate professor of medicine, was recently elected president-elect of the Association of Professors of Human and Medical Genetics (APHMG). Dr. Dasgupta, who also serves as assistant dean of admissions, will serve as president-elect from 2018-2020 and president from 2020-2022.
The mission of the APHMG is to advance education in human and medical genetics among medical students, residents, lab directors and providers. During her time in office, Dasgupta hopes to engage inter-professional communities in this mission. “I also hope to further expand the scope of our work in helping to advocate for engaging genomic medicine curricula through projects bridging the National Human Genome Research Institute (NHGRI) and the Association of American Medical Colleges,” said Dr. Dasgupta.
Dr. Dasgupta is the past chair of the Medical Genetics Course Directors Special Interest Group of the APHMG. In this position she directed national initiatives of significance for educators of medical genetics in North America. She is also an affiliate scientist of the American College of Medical Genetics, has implemented educational initiatives at the American Society of Human Genetics’ 2016 international meeting and serves on the National Institutes of Health’s (NIH) Inter-Society Coordinating Committee for Practitioner Education in Genomics and the NHGRI’s Genomic Literacy, Education and Engagement Initiative.
The Association of Professors was incorporated in 1995 to promote human and medical genetics educational programs of human and medical genetics in North American medical and graduate schools.
January 2018
2018 Wing Tat Lee Awards
Xiaoling Zhang, MD, PhD, Assistant Professor of Medicine in Biomedical Genetics is one of the recipients of the 2018 Wing Tat Lee awards, funded to establish cooperative research programs between BUSM and Chinese universities (with a preference for Hong Kong).
 Dr Zhang will examine how disease severity in Alzheimer’s Disease (AD) correlates with retinal and brain neurodegeneration. She will work with Zhigang Fan, MD, PhD, Sun Yat-sen University, to identify gene biomarkers and imaging markers in patients with mild cognitive impairment and AD to improve early diagnosis of AD. Dr. Zhang received her MD from Hubei University of Chinese Medicine and her PhD from Boston University.
Dr Zhang will examine how disease severity in Alzheimer’s Disease (AD) correlates with retinal and brain neurodegeneration. She will work with Zhigang Fan, MD, PhD, Sun Yat-sen University, to identify gene biomarkers and imaging markers in patients with mild cognitive impairment and AD to improve early diagnosis of AD. Dr. Zhang received her MD from Hubei University of Chinese Medicine and her PhD from Boston University.
* * * * * * * * *
January 2016
Fighting the Darkness
Study offers insight on genetics of age-related macular degeneration
The disease can start with blurry or fuzzy vision, or with the weird sense that straight edges—like doorframes or windowsills—actually curve. Eventually, it can lead to a total loss of sharp, central vision, leaving those affected legally blind. The disease, age-related macular degeneration (AMD), is a leading cause of blindness among the elderly worldwide, affecting more that 15 percent of people aged 65 and older, approximately 150 million people globally. More than 10 million Americans have AMD, more than those who suffer from cataracts and glaucoma combined, according to the American Macular Degeneration Foundation. While some treatments exist, there is currently no way to cure or prevent the disease.
Now, a sweeping study has identified 52 common and rare genetic variants distributed across 34 genomic regions—including 13 previously unknown—that play a role in AMD. The findings, published online in the December 21, 2015, issue of Nature Genetics, offer clues to the onset and progression of the disease that may eventually lead to better treatments or a cure. “We think of these variants as potential targets for new drug development, or biomarkers that could identify people who are at very high risk, so we could potentially intervene early, even before the disease becomes apparent,” says Lindsay A. Farrer, chief of the biomedical genetics section at Boston University School of Medicine (MED) and a co-leader of the study. “That’s the hope.”
*Please click on the title above for the full article.
* * * * * * * * *
Discovery of a novel metastasis suppressor gene in breast cancer.
Metastatic dissemination of cancer cells represents a significant clinical obstacle to curative therapy and it is the major cause of death of patients affected by cancers. Despite the advent of advanced technologies, the discovery of new metastasis suppressor genes that prevent the spread of breast cancer to distal sites using genomic efforts has been slow potentially due to their primary mode of regulation by epigenetic mechanisms. The team led by Dr. Sam Thiagalingam report that the serum deprivation response (SDPR) is a novel metastasis suppressor gene localized to 2q32-33, a region associated with significant loss of heterozygosity in breast cancer. SDPR is regulated by epigenetic rather than genetic mechanisms enabling the cancer cells to readily adapt to new microenvironments at distal sites away from the initial sites at which the cancer cells form. Interestingly, analysis of gene expression data sets indicated that loss of SDPR occurs in multiple tumor types including breast, bladder, colorectal, kidney, lung, pancreatic and ovarian cancers as well as in sarcomas. Follow up studies of this research may help with better prognosis and in the development of precision cancer therapies of metastatic cancer.
Ozturk S, Papageorgis P, Lambert AW, Wong CK, Thiagalingam A, Abdolmaleky HM, Cohen HT, and Thiagalingam S, 2016. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. SDPR functions as a metastasis suppressor in breast cancer by promoting apoptosis. Proc. Natl. Acad. Sci. USA. 113(3): 638-643. http://www.pnas.org/content/113/3/638.full.pdf?with-ds=yes
Editors’ choice: Science 22 Jan 2016:Vol. 351, Issue 6271, pp. 351. http://science.sciencemag.org/content/351/6271/twil
http://medicalxpress.com/news/2016-01-epigenetic-metastatic-breast-cancer-prognosis.html
* * * * * * * * *
November 2015
New breast cancer stem cell clues may help develop therapeutics Basal-like breast cancer is an aggressive subtype of breast cancer that is often enriched with cancer stem cells, which may drive metastasis, resistance to chemotherapy, and disease recurrence. The team led by Dr. Sam Thiagalingam found that periostin, a matricellular protein, secreted by many basal-like breast cancer cells regulates key cytokines that maintain the stem cell state. This work suggests that tumor cell-derived factors may allow basal-like breast cancer cells to establish their own functional niche, which acts to maintain a population of cancer stem cells.
Stem Cell News: http://tinyurl.com/zhzxykr
EurekaAlert: http://tinyurl.com/njj3w7z
The Daily Free Press: http://tinyurl.com/jle2qzy
Oncology Central: http://tinyurl.com/hwuld7d
Specialty Pharmacy Times: http://tinyurl.com/grhk2jw
Lambert AW, Wong CK, Ozturk S, Papageorgis P, Raghunathan R, Alekseyev YO, Gower A, Reinhard BM, Abdolmaleky HM, and Thiagalingam S. 2016. Tumor cell derived periostin regulates cytokines that maintain breast cancer stem cells. Mol. Cancer Res. 14:1 103-113. http://mcr.aacrjournals.org/content/14/1/103.long; Highlights: http://mcr.aacrjournals.org/content/14/1/1.full.pdf+html
* * * * * * * * *
Panos Papageorgis, Ph.D. receives Cyprus Research Award – Young Researcher 2015 in Life Sciences
Dr Panagiotis Papageorgis, Assistant Professor of Molecular Biology at the European University Cyprus has been awarded the prestigious “Cyprus Research Award – Young Researcher 2015” of the Research Promotion Foundation of the Republic of Cyprus in the thematic section “Life Sciences”. He received this award from the Minister of Finance Mr. Harris Georgiades during a special ceremony held on Monday Nov 23rd, 2015 at the Hilton hotel Cyprus, Nicosia. Dr. Papageorgis was formerly a Genetics and Genomics graduate student (2004-2009) in the Biomedical Genetics Section with Dr. Sam Thiagalingam and he continued to work as a joint-post doctoral fellow with him on breast cancer metastasis (2010-2014).
*Please click on the title above for the full article.
* * * * * * * * *
August 2015
Dr. Sam Thiagalingam is a recipient of a pilot grant from the BU Clinical and Translational Science Institute and the Department of Medicine. The award was provided for the grant entitled “Targeting IL13Rα2 to suppress basal-like breast cancer metastasis”. The goal of the proposed studies is to examine the clinical utility of IL13Rα2 as prognostic biomarker and to decipher novel therapeutic targets. This work is a follow up recent studies reported in Breast Cancer Research by the Thiagalingam (BUSM) and the Constantinou (University of Cyprus) laboratories (see http://www.bumc.bu.edu/busm/2015/08/05/novel-prognostic-marker-and-potential-therapeutic-target-for-triple-negative-breast-cancer-identified/)
A novel prognostic biomarker and therapeutic target for triple negative breast cancer identified
– Basal-like breast cancer (BLBC), commonly known as the triple negative breast cancer is among the most aggressive tumors with a prevalence of 15-20% of all cases of breast cancer irrespective of the ethnicity or method of analysis. A cell surface receptor, IL13Rα2 (IL13R alpha2) abundant in metastatic or late-stage BLBC has been identified by a team led by Dr. Sam Thiagalingam at the Biomedical Genetics/Boston University School of Medicine and by Dr. Constantinou at the University Cyprus. These researchers are hopeful that their discovery may lead to breakthroughs in diagnosis and therapy of breast cancer.
Please click on the following links for more information:
Boston University School of Medicine: http://tinyurl.com/pflox4o
Consumer Affairs: http://bit.ly/1IewVGW
NewsMax Health: http://ow.ly/QmDRX
Science World Report: http://tinyurl.com/o5pjb5o
Mammary Cell News: http://tinyurl.com/nevhv3q
OncoTherapy Network: http://tinyurl.com/o2c9net
* * * * * * * * *
July 2015
- IGNITION Award 2015
- Life Sciences: Lindsay Farrer, PhD (BU Distinguished Professor of Genetics) will generate pilot data on plexin-A4 isoform levels in brain, which may serve as a therapeutic target and development of a biomarker assay for Alzheimer disease.
- Spivack Award 2015 – Lindsay Farrer, Ph.D., is the recipient of the Jack Spivack Excellence in Neurosciences Awards for 2015.
– Mr. Spivack established this award in 2013 to recognize and support the research of an outstanding BUSM faculty conducting either clinical or basic research in Parkinson’s, Alzheimer’s, Chronic Traumatic Encephalopathy and other neurological disorders. Dr. Farrer is a BU Distinguished Professor of Genetics, Professor of Medicine, Neurology and Ophthalmology, and Chief of the Biomedical Genetics Section of the Department of Medicine. He also is a professor of Epidemiology and Biostatistics at the BU School of Public Health and directs the BU Transformative Training Program in Addiction Science and the BU Molecular Genetics Core Facility. Dr. Farrer is a Founding Fellow of the American College of Medical Genetics (ACMG) and a project director for the Multi-Institutional Research in Alzheimer’s Genetic Epidemiology (MIRAGE) study. He co-directs analysis for the Alzheimer Disease Genetics Consortium, and serves on the executive committee of the Alzheimer Disease Sequencing Project. He has more than 350 publications on genetic risk factors for several neurodegenerative and other chronic diseases. In collaboration with laboratories worldwide, Dr. Farrer has identified genes causing complex disorders including Alzheimer disease, age-related macular degeneration, and substance dependence. – BUSM Alumni Magazine – Spivack Award 2015
- Kenneth H. Albrecht, Ph.D., has been appointed as Director of Transgenic Core Facility.
* * * * * * * * *
May 2015
- Lindsay A. Farrer, Ph.D., has been named the BU Distinguished Professor of Genetics. Dr. Farrer also is Professor and Chief of the Biomedical Genetics Section in the department of Medicine and Professor of Neurology and Ophthalmology at BUSM, as well as Professor of Epidemiology and Biostatistics, School of Public Health, Boston University. Since 1998, he has been Director of the Boston University Molecular Genetics Core Facility.
- Dr. Sam Thiagalingam received one of the two seed grants for 2015 awarded by the Genome Science Institute (GSI). The support received from the seed grant will enable Dr. Thiagalingam to investigate, “Therapeutic targeting of SMAD4-defective drug resistant metastatic colon cancer”. The study proposes to develop a strategy using model colon cancer cells, to identify the molecular basis for resistance to common chemotherapeutic agents in metastatic colorectal cancer defective for SMAD4 and identify gene targets for potential combination therapy.
- “Systems Biology of Cancer” edited by Sam Thiagalingam, Ph.D. A book edited by Dr. Sam Thiagalingam entitled “Systems Biology of Cancer” was released by the publisher Cambridge University Press on April 9, 2015. It provides a comprehensive account of the current state of the knowledge and a road map to formulate a blueprint for the biological basis of cancer. The emphasis of the book is on the biology with minimal mathematical derivations and computational methods which are still evolving to meet the unprecedented amount of data produced with high-throughput experimentation methods that are continuing to be developed. Readers will be able to get a realistic view of what is known and remaining to be uncovered in the chapters presented by international experts on the aberrations in the fundamental biological processes, deregulation of major signaling networks, alterations in major cancers and the strategies for using the scientific knowledge for effective diagnosis, prognosis and drug discovery to improve public health. This will serve as a useful text on cancer suitable for students and researchers in all stages of their careers including the undergraduates. The contents are most suitable and highly recommended for graduate students, medical residents and fellows and experienced researchers. This landmark volume consists of a collection of 32 chapters with contributions from more than 75 international experts which includes 8 chapters by Boston University faculty and trainees. At this time the publisher is providing a 20% discount (see Flyer). Thiagalingam-Flyer-SystemsBiologyof Cancer-2015
* * * * * * * * *
November 2014
Special Contribution Teaching Award to
Shoumita Dasgupta, PhD http://www.bumc.bu.edu/medicine/files/2010/03/November-2014-newsletter1.pdf
* * * * * * * * *
August 10, 2014
From Seder to Yoga, a Jewish Approach to Treating Alzheimer’s Disease: http://forward.com/culture/203711/from-seder-to-yoga-a-jewish-approach-to-treating-a/
* * * * * * * * *
August 4, 2014
FOR IMMEDIATE RELEASE
Contact: Gina DiGravio, 617-638-8480, gina.digravio@bmc.org
RESEARCHERS IDENTIFY POTENTIAL GENE THAT MAY INCREASE RISK OF ALZHEIMER’S DISEASE IN AFRICAN AMERICANS
(Boston)– Researchers from Boston University School of Medicine (BUSM) report that two rare variants in the AKAP9 gene significantly increase the risk of Alzheimer’s disease (AD) in African-Americans. This previously unknown association furthers the understanding of the role of genetic factors in the development of AD, according to the researchers, whose findings appear in Alzheimer’s & Dementia. AD is the most frequent age-related dementia affecting 5.4 million Americans including 13 percent of people age 65 and older and more than 40 percent of people age 85 and older. Up to 75 percent of AD cases are thought to have a genetic basis; however the specific genes involved likely differ between ethnic populations. The most well-known AD risk gene, APOE4, does not play as strong a role in AD risk in African Americans as it does in Caucasians, despite the fact that a higher proportion of African Americans than Caucasians are afflicted with this disorder. By analyzing the DNA sequence for all genes from participants of the Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) Study and Genetic and Environmental Risk Factors for Alzheimer’s Disease among African Americans (GenerAAtions) Study, researchers identified two genetic variants in AKAP9 unique to African Americans that are enriched in individuals with AD. They then confirmed this association in several thousand other African American subjects in the Alzheimer Disease Genetics Consortium dataset. Carriers of either of these AKAP9 variants have a respective 2.8 and 3.6 times greater risk of developing AD. According to the researchers AKAP9 encodes a protein with multiple forms, One of these, AKAP450, is expressed in the brain and responsible for microtubule anchoring and organization. Another protein, tau, which is responsible for microtubule functioning is well known to be the key constituent of neurofibrillary tangles that accumulate in AD brains. “While further work is needed to clarify the causal link between these AKAP9 variants and AD, “this study indicates a new potential disease mechanism in the quest for a better understanding of AD, particularly in African Americans,” explained senior author Lindsay Farrer, PhD, Chief of Biomedical Genetics and professor of medicine, neurology, ophthalmology, epidemiology and biostatistics at BUSM. “Moreover, this is the first authentic example of rare genetic variants conferring a high risk of AD in African Americans,” he added. Funding for this study was provided by the National Institute on Aging (R01-AG025259, R01-AG09029, P30-AG13846, and U01-032984)
* * * * * * * * * *
July 28, 2014
FOR IMMEDIATE RELEASE
Contact: Gina DiGravio, 617-638-8480, gina.digravio@bmc.org
Researchers Identify Potential Biomarker for AD
(Boston)– Researchers from Boston University School of Medicine (BUSM) report variants in a new gene, PLXNA4, which may increase the risk of developing Alzheimer’s disease (AD). The discovery of this novel genetic association may lead to new drug treatment options that target PLXNA4 specifically. These findings appear in the Annals of Neurology. AD is the most frequent age-related dementia affecting 5.4 million Americans including 13 percent of people age 65 and older, and more than 40 percent of people age 85 and older. Genetic factors account for much of the risk for developing AD with heritability estimates between 60 percent and 80 percent. However much of the genetic basis for the disease is unexplained. Less than 50 percent of the genetic contribution to AD is supported by known common genetic variations. Using data from the Framingham Heart Study, the researchers obtained strong evidence of an association with several single nucleotide polymorphism in PLXNA4, a gene which had not been previously linked to AD. They then confirmed this finding in a larger dataset from the Alzheimer’s Disease Genetics Consortium and other datasets. Next, they performed a series of experiments in models that pinpointed the mechanism by which this gene affects AD risk. “Importantly, this is one of few single studies which go from gene finding to mechanism,” explained corresponding author Lindsay Farrer, PhD, Chief of Biomedical Genetics and professor of medicine, neurology, ophthalmology, epidemiology and biostatistics at BUSM. According to the researchers a form of the protein encoded by this gene promotes formation of neurofibrillary tangles consisting of decomposed tau protein, one of the two pathological hallmarks of the disease. “We showed that PLXNA4 affects the processing of tau as it relates to neurofibrillary tangles, the primary marker of AD. Most drugs that have been developed or that are in development for treating AD are intended to reduce the toxic form of beta-amyloid, a sticky substance that accumulates in the brain of persons with AD, and none have been very effective. Only a few drugs have targeted the tau pathway,” added Farrer. This study was supported by grants from the National Institute on Aging (R01-AG025259, P30-AG13846, R01-AG0001, U24-AG021886, U24-AG26395, R01-AG041797 and P50-AG005138), the Alzheimer Association, the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (#A110742), and the Evans Center for Interdisciplinary Biomedical Research (ECIBR) ARC on “Protein Trafficking and Neurodegenerative Disease” at Boston University.
* * * * * * * * * *
July 7, 2014
FOR IMMEDIATE RELEASE
Contact: Gina DiGravio, 617-638-8480, gina.digravio@bmc.org
Boston University Researchers and Collaborators Receive $12.6 Million NIH Grant to Study Genetics of Alzheimer’s Disease
(Boston) – Researchers from the Biomedical Genetics division of the Boston University School of Medicine (BUSM) are part of a five-university collaboration receiving a $12.6 million, four-year grant from the National Institute on Aging (NIA), part of the National Institutes of Health (NIH), to identify rare genetic variants that may either protect against, or contribute to Alzheimer’s disease risk. At BUSM, the Consortium for Alzheimer’s Sequence Analysis (CASA) is led by Lindsay A. Farrer, PhD, Chief of Biomedical Genetics and professor of medicine, neurology, ophthalmology, epidemiology, and biostatistics, who is the principal investigator. Other Boston University investigators include Kathryn Lunetta, PhD, professor of biostatistics; Gyungah Jun, PhD, assistant professor of medicine, ophthalmology and biostatistics; and Richard Sherva, PhD, research assistant professor of medicine. CASA investigators will analyze whole exome and whole genome sequence data generated during the first phase of the NIH Alzheimer’s Disease Sequencing Program, an innovative collaboration that began in 2012 between NIA and the National Human Genome Research Institute (NHGRI), also part of NIH. They will analyze data from 6,000 volunteers with Alzheimer’s disease and 5,000 older individuals who do not have the disease. In addition, they will study genomic data from 111 large families with multiple members who have Alzheimer’s disease, mostly of Caucasian and Caribbean Hispanic descent to identify rare genetic variants. “This is an exciting opportunity to apply new genomic technologies and computational methods to improve our understanding of the biological pathways underlying this disease,” said Farrer. “The genes and pathways we identify as integral to the Alzheimer process may become novel therapeutic targets,” he added. Alzheimer’s disease, a progressive neurodegenerative disorder, has become an epidemic that currently affects as many as five million people age 65 and older in the United States, with economic costs that are comparable to, if not greater than, caring for those of heart disease or cancer. Available drugs only marginally affect disease severity and progression. While there is no way to prevent this disease, the discovery of genetic risk factors for Alzheimer’s is bringing researchers closer to learning how the genes work together and may help identify the most effective interventions. This effort is critical to accomplishing the genetic research goals outlined in the National Plan to Address Alzheimer’s Disease, first announced by the U.S. Department of Health and Human Services in May 2012 and updated annually. Developed under the National Alzheimer’s Project Act, the plan provides a framework for a coordinated and concentrated effort in research, care, and services for Alzheimer’s and related dementias. Its primary research goal is to prevent and effectively treat Alzheimer’s disease by 2025. With the current award, CASA joins the NHGRI Large-Scale Sequencing and Analysis Centers program, an NIH-supported consortium that provides large-scale sequence datasets and analysis to the biomedical community. CASA researchers will facilitate the analyses of all Alzheimer’s Disease Sequencing Project (ADSP) and additional non-ADSP sequence data to detect protective and risk variants for Alzheimer’s disease. “We are delighted to support the important research being accomplished under this broad-based, collaborative effort. A team effort is vital to advancing a deeper understanding of the genetic variants involved in this complex and devastating disease and to the shared goal of finding targets for effective interventions,” said NIH Director Francis Collins, MD, PhD. “Alzheimer’s disease research is appropriately one of our highest priorities,” said BUSM Dean Karen Antman, MD “We need more to better understand the genetic and environmental mechanisms that will come in part from CASA to develop more effective treatments or even better, to prevent the disease,” she added. CASA is a collaboration of Boston University School of Medicine and four other American universities. Jonathan Haines, PhD, will lead the project at Case Western Reserve University; Richard Mayeux, MD, at Columbia University; Margaret Pericak-Vance, PhD, at the University of Miami; Gerard D. Schellenberg, PhD, at the University of Pennsylvania; and Lindsay Farrar, PhD, at Boston University. This research is supported by the NIA grant UF1-AG047133. Originally established in 1848 as the New England Female Medical College, and incorporated into Boston University in 1873, Boston University School of Medicine today is a leading academic medical center with an enrollment of more than 700 medical students and more than 800 masters and PhD students. Its 1,246 full and part-time faculty members generated more than $335 million in funding in the 2009-2010 academic year for research in amyloidosis, arthritis, cardiovascular disease, cancer, infectious disease, pulmonary disease and dermatology among others. The School is affiliated with Boston Medical Center, its principal teaching hospital, the Boston and Bedford Veterans Administration Medical Centers and 16 other regional hospitals as well as the Boston HealthNet.
* * * * * * * * * *
March 7, 2014
GENOME-WIDE STUDIES IMPLICATE CALCIUM SIGNALING AND NEURONAL POTASSIUM CHANNEL FUNCTION GENES IN ADDICTION TO COCAINE AND OPIATES
Dr. Lindsay Farrer, Professor and Chief of the Biomedical Genetics division and Director of TTPAS, and his colleagues reported in several recently published papers the discovery of multiple genes associated with risk of addiction to cocaine, opioids, and alcohol. Although it has been recognized for several decades that there is a strong heritable component of addiction to many drugs, robust evidence for contribution of specific genes has been meager, especially for dependence on cocaine or opioids. Dr. Farrer has been studying genetic factors predisposing to substance dependence for more than 15 years in collaboration with other researchers at
Figure 1: Association results from the KCNG2 gene on chromosome 18 with opioid dependence 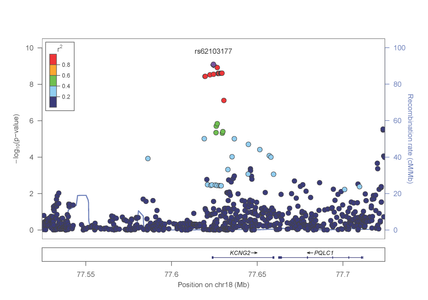
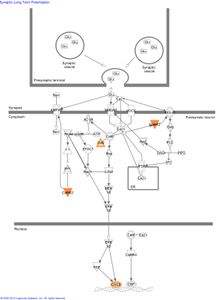
Several genes with the strongest evidence for association are clustered in biological pathways involved in the regulation of calcium and potassium levels and synaptic long term potentiation in neurons (figure 2).
Figure 2: The role of genes identified in GWAS of AA subjects in the canonical pathway “synaptic long term potentiation.”
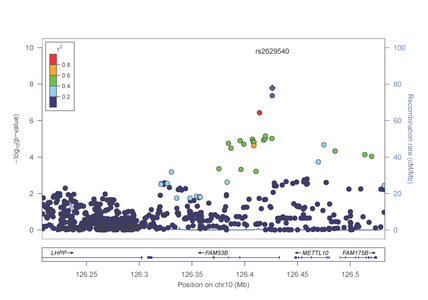
In a separate paper published in Molecular Psychiatry (http://www.nature.com/mp/journal/vaop/ncurrent/pdf/mp201399a.pdf), the investigative team reported a genome-wide significant association between risk for cocaine dependence and a SNP in FAM53B, a gene involved in regulating cell proliferation (figure 3).
Figure 3: Association results from the FAM53B gene on chromosome 10 with cocaine dependence.
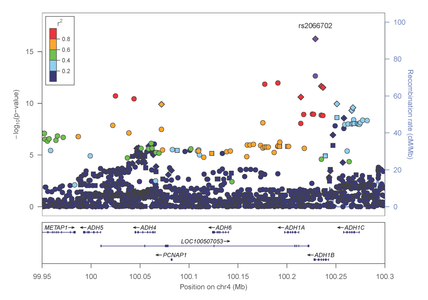
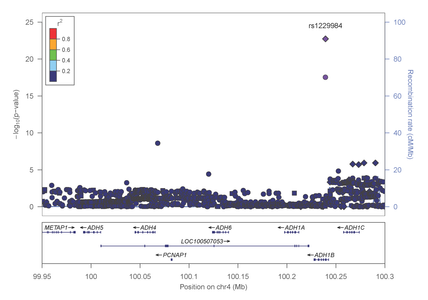
This finding was evident in both AA and EA samples. Intriguingly, several of the most significantly associated genes cluster in pathways related to calcium/potassium signaling. Most recently, Dr. Farrer, Dr. Sherva and their collaborators, identified association of alcohol dependence with several previously reported mutations in alcohol metabolizing enzymes. Importantly, the most significant findings were observed with two distinct non-synonymous (.i.e., predicted to be functionally relevant) mutations in the alcohol dehydrogenase 1B gene (ADH1B) that are population-specific (figures 4 and 5).
Figure 4: Association results within the alcohol metabolizing gene cluster on chromosome 4 in AAs
Figure 5: Association results within the alcohol metabolizing gene cluster on chromosome 4 in EAs
Novel associations were also identified in one small intergenic region on chromosome 2 in both the EA and AA cohorts, and population-specific significant associations were identified with markers located on chromosomes 5, 9 and 19. These findings were also published in Molecular Psychiatry (http://www.nature.com/mp/journal/vaop/ncurrent/full/mp2013145a.html) These results reveal several interesting facts about addiction. First, certain risk variants are population-specific while others are shared among individuals of AA and EA ancestry. Second, the genetic susceptibility to substance dependence is likely mediated by proteins involved in both the metabolism of substances as well as those involved in brain development, learning, and neuronal excitability. Finally, some genes appear to alter an individual’s risk for dependence on a specific substance, while others act as determinants of general substance dependence susceptibility.
____________________________________________________________________________________________
October 27, 2013
Eleven New Genetic Susceptibility Factors for Alzheimer’s Disease Discovered Through the Largest Study Lindsay Farrer, chief of Biomedical Genetics at Boston University School of Medicine, is interviewed on exciting new Alzheimer Research results from the International Genomics Alzheimer’s Project in an article by Science Daily.
September 10, 2013
Boston Babies to Have DNA Sequenced in Genome Study (Boston) –
Lindsay Farrer, chief of Biomedical Genetics at Boston University School of Medicine, was highlighted in an article by The Daily Free Press about a study that will have the DNA of many Boston babies sequenced. Dr.Farrer gives his insight about the five-year study that will explore the effect of DNA sequencing on the future medical care of Boston newborns.
July 2013 Genomic Studies of Non-Caucasians Could Deepen Understanding of Alzheimer’s Disease
(Boston) Neurology Reviews interviews Lindsay Farrer, chief of Biomedical Genetics at Boston University School of Medicine, regarding his presentation on Alzheimer Research of non-Caucasions at the Alzheimer’s Association International Conference 2013.
Spring 2013
Biomedical Genetics in Spring 2013 BUMC Campus Alumni News
(Boston) – Four researchers and one educator from the BUMC Biomedical Genetics section were highlighted in the Spring 2013 edition of the Boston University School of Medicine Campus Alumni News. Shoumita Dasgupta, Ph.D., is quoted in a section of the front page article, Creating Dr.Right, describing the interdisciplinary training of next generation biomedical scientists. According to the article covering the $2.5 Million Addiction Training Grant Awarded by Burroughs Wellcome Fund, Lindsay Farrer, Ph.D., and Timothy Heeren, Ph.D., professor of biostatistics at Boston University School of Public Health (BUSPH), will lead the Transformative Training Program in Addiction Science (TTPAS). Clinton Baldwin, Ph.D., and Mark Logue, Ph.D., are co-first authors of a paper detailing the findings of a newly identified gene linked to PTSD. Gyungah Jun, Ph.D., was mentioned as the lead author for a paper about the study that identified a gene linking cataracts and Alzheimer’s disease.
March 3, 2013
International consortium discovers seven new genomic regions associated with age-related macular degeneration
(Boston) – An international group of researchers has discovered seven new regions of the human genome—called loci—that are associated with increased risk of age-related macular degeneration (AMD), a leading cause of blindness. The AMD Gene Consortium, a network of international investigators representing 18 research groups, also confirmed 12 loci identified in previous studies. The study, which is published online in Nature Genetics and represents the most comprehensive genome-wide analysis of genetic variations associated with AMD, was supported by the National Eye Institute (NEI), a part of the National Institutes of Health. Lindsay A. Farrer, PhD, chief of the biomedical genetics section and professor at Boston University Schools of Medicine (BUSM) and Public Health (BUSPH), is co-lead author of the study. “This compelling analysis by the AMD Gene Consortium demonstrates the enormous value of effective collaboration,” said NEI Director Paul A. Sieving, MD, PhD. “Combining data from multiple studies, this international effort provides insight into the molecular basis of AMD, which will help researchers search for causes of the disease and will inform future development of new diagnostic and treatment strategies.” Since the 2005 discovery that certain variations in the gene for complement factor H—a component of the immune system—are associated with major risk for AMD, research groups around the world have conducted genome-wide association studies to identify other loci that affect AMD risk. These studies were made possible by tools developed through the Human Genome Project, which mapped human genes, and related projects, such the International HapMap Project, which identified common patterns of genetic variation within the human genome. The AMD Gene Consortium combined data from 18 research groups to increase the power of prior analyses. The current analysis identified seven new loci near genes. As with the previously discovered 12 loci, these seven loci are scattered throughout the genome on many different chromosomes. “A large number of samples was needed to detect additional genetic variants that have small but significant influences on a person’s disease risk,” said Hemin Chin, PhD, NEI associate director for ophthalmic genetics, who assembled the consortium and helped coordinate the study. “By cataloging genetic variations associated with AMD, scientists are better equipped to target corresponding biological pathways and study how they might interact and change with age or other factors, such as smoking.” The consortium’s analysis included data from more than 17,100 people with the most advanced and severe forms of AMD, which were compared to data from more than 60,000 people without AMD. The 19 loci that were found to be associated with AMD implicate a variety of biological functions, including regulation of the immune system, maintenance of cellular structure, growth and permeability of blood vessels, lipid metabolism and atherosclerosis. As with other common diseases, such as type 2 diabetes, an individual person’s risk for getting AMD is likely determined not by one but many genes.
Further comprehensive DNA analysis of the areas around the 19 loci identified by the AMD Gene Consortium could turn up undiscovered rare genetic variants with a disproportionately large effect on AMD risk. Discovery of such genes could greatly advance scientists’ understanding of AMD pathogenesis and their quest for more effective treatments. AMD affects the macula, a region of the retina responsible for central vision. The retina is the layer of light-sensitive tissue in the back of the eye that houses rod and cone photoreceptor cells. Compared with the rest of the retina, the macula is especially dense with cone photoreceptors and is what humans rely on for tasks that require sharp vision, such as reading, driving and recognizing faces. As AMD progresses, such tasks become more difficult and eventually impossible. Some kinds of AMD are treatable if detected early, but no cure exists. An estimated 2 million Americans have AMD. Scientists have shown that age, diet, and smoking influence a person’s risk of developing AMD. Genetics also plays a strong role. AMD often runs in families and is more common among certain ethnicities, such as Asians and people of European descent. AMD typically presents later in life, but identifying genetic variants associated with the disease, all of which are present at birth, could help future studies determine how to stop the disease from progressing and even from occurring. “Genetic research allows us to piece together disease pathways that may have their starting point much earlier in life,” said Farrer. “These newly identified genes, individually and collectively, provide novel clues and targets to evaluate for their potential therapeutic benefits.”
For more information about AMD, visit http://www.nei.nih.gov/health/maculardegen/index.asp. Goncalo Abecasis, DPhil, from the University of Michigan; Iris Heid, PhD, from the University of Regensburg, Germany; and Jonathan L. Haines, PhD, from Vanderbilt University are the study’s other co-lead authors. Funding for the research conducted at BUSM for this study was provided in part by the National Institutes of Health under grant award number R01-EY014458 and the Edward N. & Della L. Thome Memorial Foundation.
March 2011
Alzheimer’s Disease Consortium Identifies Four New Genes For Alzheimer’s Disease Risk
In the largest study of its kind, researchers from a consortium led by the University of Pennsylvania School of Medicine, Boston University School of Medicine and the University of Miami identified four new genes linked to Alzheimer’s disease. Each gene individually adds to the risk of having this common form of dementia later in life. These new genes offer a portal into what causes Alzheimer’s disease and is a major advance in the field. The study, conducted by the Alzheimer’s Disease Genetics Consortium, reports genetic analysis of more than 11,000 people with Alzheimer’s disease and a nearly equal number of elderly people who have no symptoms of dementia. Three other consortia contributed confirming data from additional people, bringing the total number of people analyzed to over 54,000. The consortium also contributed to the identification of a fifth gene reported by other groups of investigators from the United States, the United Kingdom, France and other European countries. The findings appear in the current issue of Nature Genetics. The study is the result of a large collaborative effort with investigators from 44 universities and research institutions in the United States, led by Gerard D. Schellenberg, PhD, at Penn, with primary analysis sites at Boston, led by Lindsay A. Farrer, PhD, and Miami, led by Margaret A. Pericak-Vance, PhD. “This is the culmination of years of work on Alzheimer’s disease by a large number of scientists, yet it is just the beginning in defining how genes influence memory and intellectual function as we age. We are all tremendously excited by our progress so far, but much remains to be done, both in understanding the genetics and in defining how these genes influence the disease process,” Schellenberg said.
Until recently, only four genes associated with late-onset Alzheimer’s have been confirmed, with the gene for apolipoprotein E-e4, APOE-e4, having the largest effect on risk. The Nature Genetics studies add another four — MS4A, CD2AP, CD33, and EPHA1 – and contribute to identifying and confirming two other genes, BIN1 and ABCA7, thereby doubling the number of genes known to contribute Alzheimer’s disease. The researchers’ ultimate aims are two fold. First, identification of new Alzheimer’s disease genes will provide major clues as to its underlying cause. Genetic studies can provide new insights into the molecules at the center of the disease. Gaining this type of understanding is critical for drug discovery since the currently available treatments are only marginally effective. “The skyrocketing prevalence and financial and societal costs of Alzheimer’s disease will soon undermine the delivery of healthcare worldwide,” said Farrer. “That gives our national research enterprise added incentive to act quickly and boldly to make new discoveries.” Second, gene discovery of the type highlighted in the Nature Genetics article will contribute to predicting who will develop Alzheimer’s disease, which will be important when preventive measures become available.
Knowing these risk genes will also help identify the first disease-initiating steps that begin in the brain long before any symptoms of memory loss or intellectual decline are apparent. This knowledge will help researchers understand the events that lead to the destruction of large parts of the brain and eventually the complete loss of cognitive abilities. Currently, Alzheimer’s genetics researchers are coming together for an even larger, similar study. The Alzheimer’s Association in the US and the Fondation Plan Alzheimer in France have funded the formation of the International Genomics of Alzheimer’s Project, whose members met for the first time in November 2010 in Paris. The research published in Nature Genetics was supported by the National Institute on Aging, part of the National Institutes of Health, which includes 29 Alzheimer’s Disease Centers, the National Alzheimer’s Coordinating Center, the NIA Genetics of Alzheimer’s Disease Data Storage Site, the NIA Late Onset Alzheimer’s Disease Family Study and the National Cell Repository for Alzheimer’s Disease. These Centers collect, store and make available to qualified researchers DNA samples, datasets containing biomedical and demographic information about participants, and genetic analysis data. The findings from the current issue of Nature Genetics can be found here.
April 2, 2009
Sam Thiagalingam’s Lab produces cover and story for Cancer Biology & Therapy
The members of Sam Thiagalingam’s lab have have created the cover graphic for the current issue of Cancer Biology & Therapy. The issue spotlights their research regarding interaction between p53 and hBub1 in response to spindle checkpoint (SAC) activation. For more information about the research discoveries and the cover image, navigate to the webpage for the current issue.
March 10, 2009
Researchers from Genetics Program discover Gene Associated with Cocaine Dependence A reasearch group including members of the BUSM Genetics Program have discovered that the MANEA gene is associated with coacaine dependence and cocaine-induced paranoia. The findings, discovered through research in collaboration with the Yale University School of Medicine and University of Connecticut School of Medicine, were published in the March issue of the Archives of General Psychiatry. More information is available here.