Zaia Lab (CBMS) – Postdoctoral Associate Position
Postdoctoral associate position
Laboratory of Joe Zaia
Department of Biochemistry- Center for Biomedical Mass Spectrometry
We are recruiting a post-doctoral associate to work on NIH supported projects in the Zaia group at Boston University.
https://www.bumc.bu.edu/biochemcellbio/profiles/joseph-zaia/
Our group uses mass spectrometry methods for studying disease processes in tissue using combined proteomics, glycomics and glycoproteomics. We have an opening for a postdoctoral scientist to work on combined proteomics and glycomics from brain tissue. The work will involve combined glycomics and proteomics to determine the roles of glycoprotein and proteoglycan expression in neurological diseases.
Competitive salary and benefits are available commensurate with experience. Fluent spoken English, a strong publication track record, and a cooperative attitude are a must for this position. The Zaia group is part of the Center for Biomedical Mass Spectrometry at Boston University. This highly collaborative group provides excellent opportunities for career development of a young Ph.D. scientist.
Candidates should have a recent Ph.D. in analytical chemistry, biochemistry, or a related biomedical field with strong experience with mass spectrometry. Glycomics and/or proteomics experience is strongly recommended.
Candidates should send curriculum vitae, cover letter, and contact information for three letters of reference to Joe Zaia (jzaia@bu.edu).
Boston University prohibits discrimination against any individual on the basis of race, color, religion, sex, age, national origin, physical or mental disability, sexual orientation, gender identity, genetic information, military service, pregnancy or pregnancy-related condition, or because of marital, parental, or veteran status.
Congratulations to all the Biochemistry Faculty members awarded Dahod Pilot Grants
Congratulations to Alla Grishok, Valentina Perissi, Bob Varelas and Cathy Costello, who were all awarded Dahod Pilot Grants.
Alla Grishok, PhD, associate professor of biochemistry, studies the oncoprotein DOT1L, which is essential for triple negative breast cancer progression. DOT1L, localized to the nucleus, activates many genes driving uncontrolled cell divisions. The Grishok lab will study how different parts of the large DOT1L protein work together to activate cancer-promoting genes. This mechanistic work may suggest new approaches to inhibit DOT1L, a promising drug target for cancer treatment.
Valentina Perissi, PhD, associate professor of biochemistry, will study the molecular mechanism of PARP1 inhibition in human breast cancer cells. Poly-ADP ribosyltransferase inhibitors (PARPi) have emerged as promising drugs for the treatment of triple negative breast cancer and metastatic breast cancer. Understanding how PARP activity is endogenously restricted in cells will assist in determining the dosage of current PARPi, their efficacy in combination with other treatments, and the design of novel inhibitors.
Bob Varelas, PhD, associate professor of biochemistry, will collaborate with Stefano Monti, PhD, associate professor of medicine and biostatistics, to define and understand tumor-stromal signaling networks in triple negative breast cancer. The study will use knowledge gained from network structure analyses of high-dimensional data obtained from human breast cancers to determine how stromal-epithelial signaling hubs influence cell plasticity and aggressive phenotypes using in vivo and ex vivo tumor models to identify vulnerable targets for therapeutic translation.
Daniel Dempsey, PhD, assistant professor of dermatology, will collaborate with William Fairfield Warren Distinguished Professor of Biochemistry Catherine Costello, PhD, and Associate Professor of Pathology & Laboratory Medicine and Ophthalmology Nader Rahimi, PhD, to develop new chemical biology tools to dissect the role of protein modifications in promoting breast cancer to precisely quantitate abnormally modified proteins and define a mechanistic purpose for these changes. Unraveling these mechanisms will identify new vulnerabilities that may be exploited with novel therapeutics to treat patients with breast cancer.

Congratulations to Joe Zaia on his SfG Award.
Congratulations to Professor Joe Zaia who was awarded the 2021 ASBMB Molecular and Cellular Proteomics Lectureship Award by the Society for Glycobiology (SfG).


CBMS Receives Massachusetts Life Science Center Grant
The Center for Biomedical Mass Spectrometry (CBMS) has been awarded a grant by the Massachusetts Life Science Center to advance the ability to characterize spike protein glycosylation as respiratory viruses evolve. The CBMS applies mass spectrometry methods to meet the emerging needs in biomedicine. As part of this award, CBMS has just installed a new Waters SELECT SERIES Cyclic IMS system. The Cyclic system represents a breakthrough technology that will facilitate rapid and accurate characterization of virus spike protein glycosylation to support virus surveillance and vaccine development.
Respiratory viruses including influenza and coronaviruses evolve rapidly as they circulate in the human population. These proteins are coated with a spike protein that recognizes receptors in host airway cells. Spike proteins are decorated by sugar molecules known as glycans. These glycans enhance the structures and functions of proteins, including the virus spike proteins. Researchers track the genetic sequences of viruses as they evolve. The genetic sequences do not predict the manner in which glycans alter the structure and function of virus spike proteins.
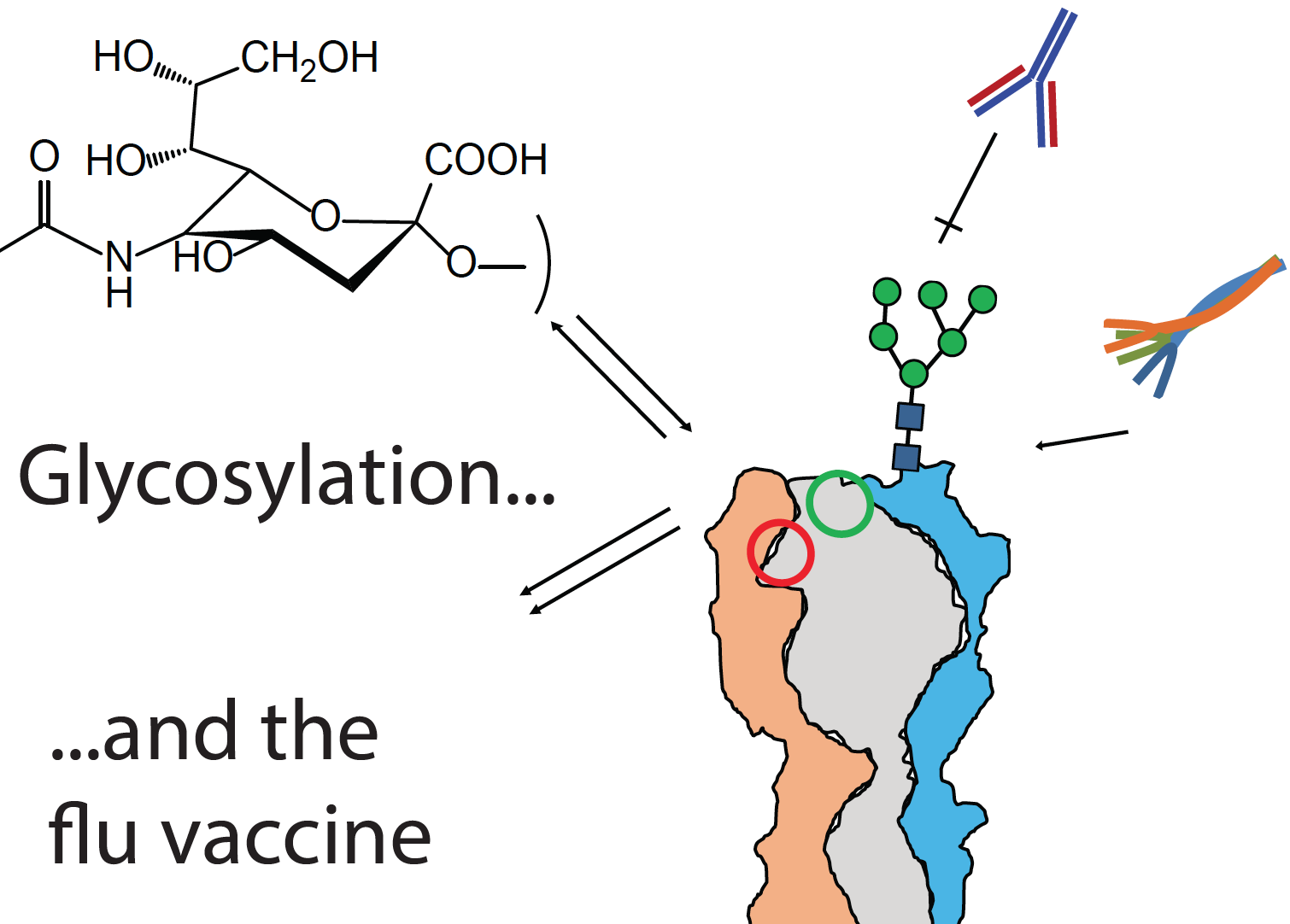
Building a better flu vaccine
New research from the Zaia laboratory has been published in Molecular Proteomics: "Why glycosylation matters in building a better flu vaccine".
Low vaccine efficacy against seasonal influenza A virus (IAV) stems from the ability of the virus to evade existing immunity while maintaining fitness. While most potent neutralizing antibodies bind antigenic sites on the globular head domain of the IAV envelope glycoprotein hemagglutinin (HA), the error-prone IAV polymerase enables rapid evolution of key antigenic sites, resulting in immune escape. Significantly, the appearance of new N-glycosylation consensus sequences (sequons, NXT/NXS, rarely NXC) on the HA globular domain occurs among the more prevalent mutations as an IAV strain undergoes antigenic drift. The appearance of new glycosylation shields underlying amino acid residues from antibody contact, tunes receptor specificity, and balances receptor avidity with virion escape, all of which help maintain viral propagation through seasonal mutations. The determination of site-specific glycosylation of IAV glycoproteins would enable development of vaccines that take advantage of glycosylation-dependent mechanisms whereby virus glycoproteins are processed by antigen presenting cells
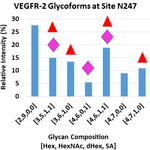
Research Discoveries: VEGFR2 glycosylation
Vascular endothelial growth factor receptor 2 (VEGFR2) is a heavily N-glycosylated pro-angiogenic receptor tyrosine kinase. Stimulation of the receptor by vascular endothelial growth factor (VEGF) induces quiescent endothelial cells to proliferate and sprout, and VEGFR2 signaling is also required for tumor growth and metastasis. In a new paper in the Journal of Biological Chemistry by Kevin Brown Chandler and colleagues in the Costello and Rahimi laboratories demonstrated that the glycosylation status of VEGFR2 alters signaling through the receptor. Specifically, sialic acid-capped N-glycans at site N247 oppose ligand-mediated receptor activation, whereas asialo-glycans (lacking sialic acid) favor VEGFR2 activation.
Russek Award Winners in Biochemistry Laboratories
The Henry I. Russek Award Nominating Committee and the Russek Executive Committee would like to announce the winners of this year’s Department of Biochemistry awards. The honorable mention was awarded to Deborah Chang who is a student in Dr. Zaia’s lab, second prize was awarded to Julia Hicks-Berthet who is a student in Dr. Varelas's lab and the first prize was awarded to Elena Stampouloglou, who is a student in Dr. Varelas's lab. There is more great news for our department! The first prize awardee for the Molecular & Translational Medicine program is Rekha Raghunathan, a student in Dr. Zaia’s lab and the first prize awardee for the Genetics & Genomics program is Stefanie Chan, a student in Dr. Perissi’s lab. Congratulations to all of the awardees (and their mentors)!
So, we'll see you all this Friday, April 26th in Hiebert Lounge for Henry I. Russek Student Achievement Day 2019 and the keynote address to be delivered by Dr. Xiowei Zhuang; her talk is entitled “Illuminating Biology at the Nanoscale and Systems Scale by Imaging”.

Dr. Costello receives HUPO 2019 Lifetime achievement award
Catherine Costello Receives Lifetime Achievement in Proteomics Award
Catherine Costello, PhD, the William Fairfield Warren Distinguished Professor at Boston University School of Medicine (BUSM), has received the 2019 Lifetime Achievement in Proteomics Award from the U.S. Human Proteome Organization (U.S. HUPO). This inaugural award recognizes a career of discovery that has made a lasting impact in the field of proteomics, the field which explores the distribution, dynamics and modifications of proteins in cells and living organisms and their relationships to health and disease. ... (link)

Dr. Cathy Costello honored with special issue in The Journal of the American Society for Mass Spectrometry

In recognition of Dr. Catherine Costello's 2017 ASMS Award for a Distinguished Contribution in Mass Spectrometry, the entire June 2018 issue focused on Mass Spectrometry in Glycobiology and Related Fields and was a tribute to Dr. Costello's numerous contributions to the field. The first article in this issue written by Joe Zaia and Veronica Bierbaum describes Dr. Costello's vision and leadership in glycoscience mass spectrometry.
Please also read about the current research in the Costello lab.
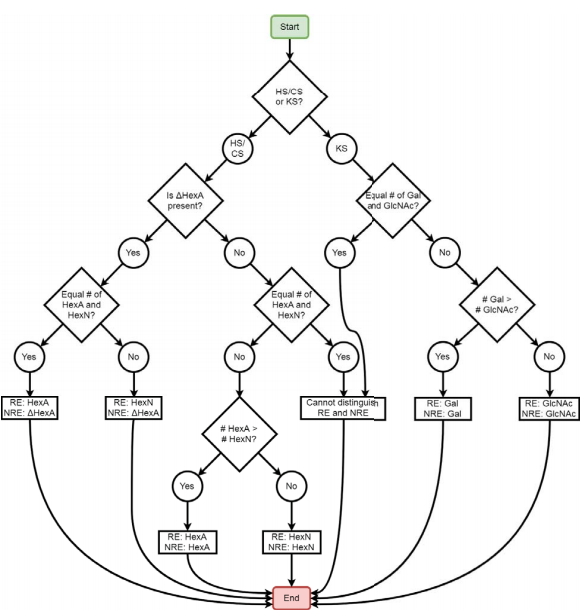
New Software Developed for Glycosaminoglycan Analysis
The Zaia laboratory's recent study: “Software for peak finding and elemental composition assignment for glycosaminoglycan tandem mass spectra” has been selected to appear in a special virtual issue of ASBMB journal content on Omics of lipids, glycans and polar metabolites. This paper was selected from hundreds of papers and best represents the exciting advances made in studying these systems over the last three years.
Glycosaminoglycans (GAGs) covalently linked to proteoglycans are characterized by repeating disaccharide units and variable sulfation patterns along the chain. The Zaia laboratory and others have demonstrated the usefulness of tandem mass spectrometry (MS2) for assigning the structures of GAG saccharides; however, manual interpretation of tandem mass spectra is time-consuming, so computational methods must be employed. The Zaia group developed GAGfinder, the first tandem mass spectrum peak finding algorithm developed specifically for GAGs. GAGfinder is a targeted, brute force approach to spectrum analysis that utilizes precursor composition information to generate all theoretical fragments. GAGfinder also performs peak isotope composition annotation, which is typically a subsequent step for averagine-based methods.