The Zaia laboratory’s recent study: “Software for peak finding and elemental composition assignment for glycosaminoglycan tandem mass spectra” has been selected to appear in a special virtual issue of ASBMB journal content on Omics of lipids, glycans and polar metabolites. This paper was selected from hundreds of papers and best represents the exciting advances made in studying these systems over the last three years.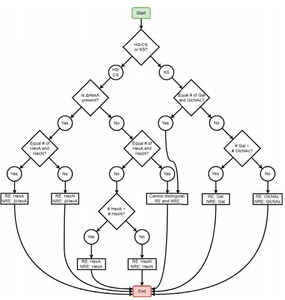
Glycosaminoglycans (GAGs) covalently linked to proteoglycans are characterized by repeating disaccharide units and variable sulfation patterns along the chain. The Zaia laboratory and others have demonstrated the usefulness of tandem mass spectrometry (MS2) for assigning the structures of GAG saccharides; however, manual interpretation of tandem mass spectra is time-consuming, so computational methods must be employed. The Zaia group developed GAGfinder, the first tandem mass spectrum peak finding algorithm developed specifically for GAGs. GAGfinder is a targeted, brute force approach to spectrum analysis that utilizes precursor composition information to generate all theoretical fragments. GAGfinder also performs peak isotope composition annotation, which is typically a subsequent step for averagine-based methods.