2023 Early Career Development Awards
Last year we introduced a pilot grants program meant to enhance the careers of our postdoctoral trainees.
Again in 2023, we will award two, one-year pilot grants of $20,000 each to postdoctoral trainees for projects based on an innovative idea that establishes a new research direction. Each award will include a $1,000 supplement to the salary of the applicant. These grants are intended to provide trainees with the experience of developing a fundable research plan that can be pursued in their current labs, or included in future job applications. The guidelines of the program are as follows:
1. Eligibility: Applicants must be Postdoctoral Associates/Fellows, Instructors, or Research Assistant Professors in the BUSM Department of Biochemistry. Preference will be given to applicants who have not received previous external research funding as a Principal Investigator.
2. The project can take the applicant’s current research as a starting point, but must represent a new and innovative direction that is developed primarily by the applicant. Projects should not merely duplicate current or planned directions of the applicant’s PI.
3. Applications should include the following: (1) A research plan of no more than three pages (including figures); (2) References (no page limit); (3) A one‐page description of the applicant’s research background, accomplishments, and career plans; and (3) A budget with no more than $10,000 of salary support for the applicant. A short justification should be included for each budget item. Documents should use 11‐point font and 1⁄2 inch margins. Applicants should also include copies of their full CV.
4. A letter of support from the applicant’s PI should be included. In addition to commenting on the applicant’s qualifications, and approving the applicant’s commitment of time to the project, the letter should attest to the fact that development of the research project and preparation of the application were the work of the applicant.
Applications will be reviewed by a committee of faculty from within and outside the department, and the two selected applicants will each deliver an award lecture to the department at the end of the grant period. Feedback will be provided to all applicants by the reviewers. Interested trainees should submit a letter of intent by December 1st, including the applicant’s name, mentor’s name, and provisional title.
Full applications are due March 15th and Decisions will be announced by April 1st.
Submit Letter of Intent—Due March 1
Submit Full Application—Due March 15
Mentor—Submit Letter of Support—Due March 15
PhD Student Alejandro Rondón Ortiz Selected as Tau Leadership Fellow
“These early-career leaders in research will know their value goes beyond the bench, and that their skills and expertise can make an impact on their community and inspire future scientists” - Dr. Amy Rommel, Scientific Program Director for the Rainwater Charitable Foundation
Congratulations to Alejandro N. Rondón Ortiz, a PhD candidate in Biology-Neurobiology in both the Center for Network Systems Biology and the Laboratory of Neurodegeneration from Boston University.
The Rainwater Tau leadership fellowship is an award funded by the Rainwater Charitable Foundation. This foundation invests in early-career scientist to promote the next generation of Tau researchers/leaders. Crucial factors for the award are scientific mentorship and community outreach, which Alejandro has fulfilled by training current and future graduate and undergraduate students, especially from underrepresented identities in STEM.
Describing his research and award, Alejandro said: "For the award I used the second aim from my qualifying exam written proposal. I am using state of the art/multidisciplinary approaches to explore protein-protein interactions in neurodegeneration. I generated genetic tools that encode for chimeric proteins, and these proteins label interacting protein partners with a tag. This “tagged-protein partner” can be purified and subsequently identified by proteomics. One characteristic of these chimeric proteins is that they interact with protein aggregates and biomolecular condensates, molecular hallmarks of tauopathies. These genetic tools are predicted to work in a diversity of biological systems, including cell/neuronal cultures and brains from transgenic rodents. Finally, these uncovered pathology-associated protein networks can be cross-referenced with available datasets and propose druggable targets at early stage of the disease progression."
More about Alejandro:
He holds a PharmD degree from Universidad Catolica de Santa Maria, Peru, and an MS in Pharmacology from MCPHS University, Boston.
He is interested in neurodegenerative disorders (particularly in tauopathies). Tauopathies are a group of neurodegenerative disorders that have a common denominator: the misfolding of Tau protein (e.g. Alzheimer’s disease, progressive supranuclear palsy, cortico-basal degeneration, and others). He uses multidisciplinary approaches to explore tauopathies at the molecular level, and hopes hi findings will contribute to a better understanding of these neurodegenerative disorders.
2021 Biochemistry Department Retreat
Towards the end of September the Department of Biochemistry gathered at the Science Museum in Boston for its first annual retreat in almost two years. We are a large department, and the retreat provides an opportunity to bring people together to learn about the work taking places across different laboratories and specialties, as well as meet people from the different groups. The agenda included a 'state of the department' delivered by department chair Dr. David Harris, presentations by PIs from every floor of our building, and a delicious lunch!

Congratulations UROP students
Congratulations to the undergraduate students in the Biochemistry Department that presented at the 24th Annual Undergraduate Research Symposium.
Department that presented at the 24th Annual Undergraduate Research Symposium.
- Jessica Chen, Project Title: Identification of Small Molecule Inhibitor IGGi-11 of GIV-Gai Interaction for Cancer-Associated Signaling Complex Targeting
Mentor: Mikel Garcia-Marcos - Yongting Chen, Project Title: LC-MS/MS Analysis of Glycans Derivatized with a Reducing-end Fixed Charge
Mentor: Cheng Lin - William Dorst, Project Title: Setting Up Studies of Flamenco Mutant Flies and P-element Transposition in S2 Cells
Mentor: Nelson Lau - David Garcia, Project Title: The Localization of Golgin and P2X7 in Cell-Cell Signaling and Migration in the Context of Corneal Wound Healing in Diabetic and Normal Mice Models Mentor: Vickery Trinkaus-Randall
- Amber Liu, Project Title: Characterizing the Role of Aortic Carboxypeptidase-like Protein in Connective Tissue Disease Mentor: Matthew Layne
- Timothy Liu, Project Title: Use of Electron Microscopy to Characterize Aα-PrPC Assembly’s Structures Faculty
Mentor: David Harris - Max Lohse, Project Title: Creation of a Conditional hnRNPUL1 Knock-out in Human Cell Culture Using CRISPR/Cas9
Mentor: Michael Blower - James Lu, Project Title: The Effect of the Hippo Pathway on Multiciliated Cells in the Airway Epithelium Faculty
Mentor: Xaralabos Varelas - Elaine Park, Project Title: Reconstitution of RNAi in Zebrafish Mentor: Daniel Cifuentes
- Xiaoyu Zhang, Project Title: Improving the Accuracy of Glycan Structural Assignments with Stable Isotope Labeling
Mentor: Catherine E Costello
Congratulations Dr. Kingston!
Congratulations to Nathan (Varelas Lab) who successfully defended his thesis this past Wednesday (9/29/2021).

Best of luck in your future.
Congratulations to Mikel Garcia-Marcos on his promotion to Full Professor of Biochemistry!
Congratulations to Mikel Garcia-Marcos on his promotion to Full Professor of Biochemistry!

The Garcia-Marcos Lab investigates signal transduction mechanisms with the ultimate goal of elucidating the molecular basis of human diseases and developing novel therapeutic approaches.
Congratulations to Joe Zaia on his SfG Award.
Congratulations to Professor Joe Zaia who was awarded the 2021 ASBMB Molecular and Cellular Proteomics Lectureship Award by the Society for Glycobiology (SfG).

Congratulations Dr. Hicks-Berthet!
Congratulations to Julia (Varelas Lab) who successfully defended her thesis this past Thursday (7/15/2021).
Best of luck in your future. 
Research News: New insight into signals that limit goblet cell specification in the lung.
The Varelas Lab has published a study in Cell Reports describing important roles for the transcriptional regulators YAP and TAZ, which are key effectors of Hippo pathway signaling, in lung epithelial homeostasis.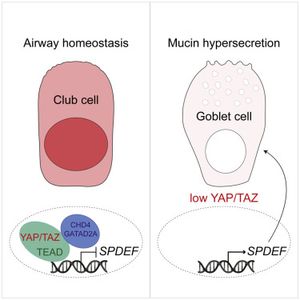
The study explored roles for YAP and TAZ in adult lung epithelium and found that conditional deletion of theYap and Wwtr1/Taz genes in animal models led to severe lung damage that included alveolar and bronchial cellular defects. One notable phenotype that was further studied was an aberrant production of mucus secreting goblet cells that resulted from the transdifferentiation of airway club cells with low YAP/TAZ activity.
Goblet cells secrete mucus that normally helps protect the lining of the bronchus and traps microorganisms for clearance. However, aberrant specification of goblet cells leads to defective lung functions that associate with a range of lung diseases (e.g. asthma, COPD, Cystic Fibrosis and chronic bronchitis). Identification of YAP/TAZ as key regulators of goblet cell production offers insight into molecular signals that may dysregulated in such diseases.
Gene expression and chromatin binding analyses from murine and human lung models revealed that YAP/TAZ repress a gene expression network that is important for controlling goblet cell production. Interestingly, altering signals that induce nuclear YAP/TAZ activity was sufficient to block goblet cell production in experimental cell models, suggesting that signals governing YAP/TAZ activity may offer a therapeutic means to restrict goblet cell production in disease.
For full text of the article, click here.
Research News: Saeed Lab Makes New SARS-CoV-2 Discoveries
The Saeed Lab, in partnership with the Broad Institute of MIT and Harvard, has published new findings in Cell.
In this study, the Saeed lab discovered SARS-CoV-2 elements being identified by human T cells to initiate an immune attack against the virus. As a background, the human immune system has two arms: innate immunity and adaptive immunity. While innate immunity launches a quick, non-specific fight against the virus, adaptive immunity is slow and utilizes specific viral components to get activated and unleash severe attack on the virus. A distinctive feature of adaptive immunity is that it “remembers” the viral identity, and if encountered by the same virus in the future, sets in motion a quick and intense campaign to overpower the virus. The adaptive immunity can be further divided into two subgroups: B cell immunity (a.k.a. humoral immunity) and T cell immunity (a.k.a. cellular immunity). While B cell immunity acts by generating antibodies against the virus, T cell immunity employs specialized cells to destroy the virus-infected cells.
Although a number of studies have reported SARS-CoV-2 elements being recognized by human B cells to produce antibodies, there remained a paucity of knowledge regarding viral components being identified by human T cells, and this is despite the fact that the role of T cells in SARS-CoV-2 clearance has been increasingly recognized. To fill this knowledge gap, the Saeed lab collaborated with a research group at the Broad Institute of MIT and Harvard and performed mass spectrometry-based analysis of viral peptides (protein fragments) that have the ability to activate T cells. This led to the discovery of 37 peptides. Strikingly, eight of these peptides originated from the hidden, non-canonical SARS-CoV-2 proteins that are often overlooked by the scientists.
The team then performed a series of experiments in human and mice to test if the identified viral peptides were capable of activating T cells. Most of them indeed were. To scientists’ surprise, some peptides derived from the hidden viral proteins provoked the strongest immune response, highlighting an unexpected yet critical role of these proteins in virus biology. These findings will change the way scientists study the human immune responses to SARS-CoV-2 and will also inform the development of next-generation vaccines. Incorporation of viral components capable of invoking strong T cell responses is expected to produce a vaccine that is more effective and broadly protective against newly emerging SARS-CoV-2 variants.
For full text of the article, click here.