Jingyi Zhao, Ph.D (Garcia-Marcos Lab) awarded 2022 Dahod International Scholar
Congratulations to Jingyi Zhao, Ph.D. (Garcia-Marcos Lab) who was awarded 2022 Dahod International Scholar.
Jingyi, a postdoc in Dr. Mikel Garcia-Marcos’ laboratory, will characterize a small molecule inhibitor of a novel signaling mechanism that promotes breast cancer metastasis. He has identified a promising candidate molecule, named IGGi-11, that disrupts a protein signaling complex specifically present in metastatic cancer cells. He intends to further confirm the specificity of this molecule in preventing cancer cell invasiveness without overt cytotoxic effects in normal cells, and to work towards developing analog compounds with improved properties in preclinical breast cancer models.
Matt Layne receives Educator of the Year Award 2022
Congratulations to Matt Layne who received Educator of the Year 2022 for the Ph.D. program. 
New Publication – Researchers Identify Novel PARP-like Enzyme in Mitochondria
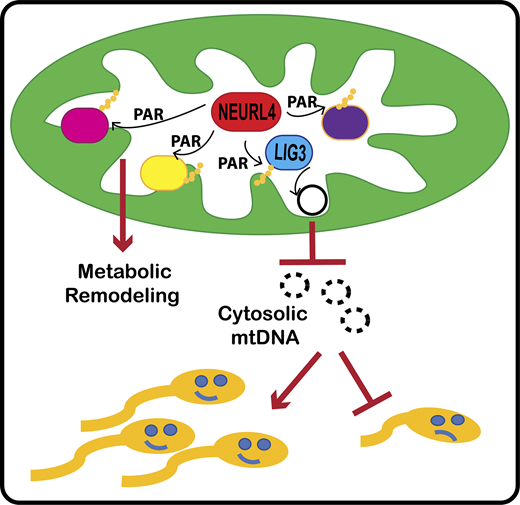
The Perissi Lab in collaboration with the Emili Lab has a new publication that came out on February 14th in the Journal of Cell Biology (JCB) "Neuralized-like protein 4 (NEURL4) mediates ADP-ribosylation of mitochondrial proteins"
For the first time, BUSM researchers have identified an ADP-ribosyltransferase enzyme that is active in the mitochondria (the organelle that generate most of the chemical energy needed to power biochemical reactions in cells) and characterized its activity. ADP-ribosyltransferases are enzymes that play a role in the modification of other proteins. The activity of this new mitochondrial enzyme, called NEURL4, is similar to that of PARP1, a nuclear enzyme well studied for its critical role in DNA damage repair and regulation of gene expression.
To characterize the enzymatic activity of NEURL4, the team led by first and co-corresponding author Maria Dafne Cardamone, PhD, former research assistant professor of biochemistry, first performed in-vitro enzymatic assays. Then they generated a cell line in which NEURL4 was deleted using a gene editing system. From there they used mass spectrometry to compare the modifications on mitochondrial proteins in wild-type (“normal”) versus cells without NEURL4.
According to the researchers this approach led them to identify a number of mitochondrial proteins as targets of NEURL4, including mitochondrial LIG3, a protein involved in the repair of damaged mitochondrial DNA and confirmed that NEURL4 activity is required for the maintenance of mitochondrial DNA (mtDNA) integrity.
While this study does not have any direct implication for clinical treatments, the team says it represents an important step in understanding how cells maintain homeostasis and respond to oxidative stress. “While this will not directly improve people’s quality of life, we hope it will provide fertile grounds for new translational applications for the treatment of diseases with a mitochondrial component, such as cancer, diabetes, age-related neurodegenerative disorders and rare mitochondrial diseases” said co-corresponding author Valentina Perissi, PhD, associate professor of biochemistry and co-director of the Boston Nutrition Obesity Research Center at BUSM.
The researchers believe future studies investigating how NEURL4-mediated modifications of specific mitochondrial proteins affect mitochondrial functions could lead to the identification of novel regulatory strategies to target for therapeutic purposes. A better understanding of NEURL4 biology may also prove relevant for the treatment of male infertility. Mitochondrial DNA deletions are linked with reduced sperm mobility and infertility in both mice and humans and this research indicates that reduced NEURL4 expression in mice is associated with increased mtDNA deletions and markers of male infertility, such as reduced sperm count and increased sperm clumping.
Andrew Emili, PhD, and Julian Kwan, PhD, from the BU Center for Network Systems Biology (CNSB) collaborated on this research.
Cathy Costello receives 2022 Wing Tat Lee Award
Congratulations to Cathy Costello who was one of six recipients of the 2022 Wing Tat Lee awards, funded to establish cooperative research programs between BUSM and Chinese universities (with a preference for Hong Kong).
Dr. Costello will use advanced analytical methods to collaborate with Huilin Li, PhD, and Yang Mao, PhD, both professors of pharmacy at Sun Yat-Sen University, Guangzhou to, (1) investigate how variations in the amino acid sequences or in post-translational modifications of key proteins affect the efficiency of entry into host cells for variants of the SARS-CoV-2 virus and (2) determine how changes in the structure of a cell surface receptor may alter the course of chronic kidney disease.
Nelson Lau receives 2022 Wing Tat Lee award
Congratulations to Nelson Lau who was one of six recipients of the 2022 Wing Tat Lee awards, funded to establish cooperative research programs between BUSM and Chinese universities (with a preference for Hong Kong).
Dr. Lau's lab will collaborate with Dr. Feifei Yin, associate professor of biology at the Hainan Medical University to characterize virus small RNAs in mosquitoes and ticks from Southern China. This collaboration will build a new BUSM Tick Small RNA Genomics resource to complement the Mosquito Small RNA Genomics resource that the Lau lab has built in collaboration with NEIDL investigators.
New Publication in Molecular & Cellular Proteomics from the Zaia Lab
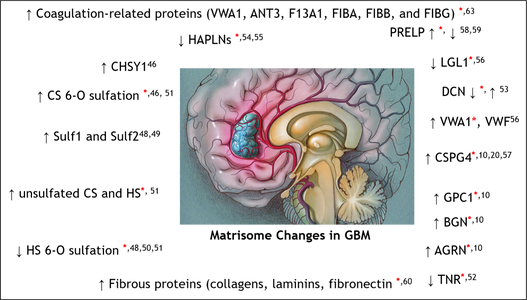
The Zaia Lab, in collaboration with Prof. Joanna Phillips of the Dept. of Nerological Surgery at the University of California San Francisco, has a new publication out in Molecular & Cellular Proteomics, "In-depth matrisome and glycoproteomic analysis of human brain glioblastoma versus control tissue."
Glioblastoma (GBM) is the most common and malignant primary brain tumor. The extracellular matrix (ECM), also known as the matrisome, helps determine glioma invasion, adhesion, and growth. Little attention, however, has been paid to glycosylation of the ECM components that constitute the majority of glycosylated protein mass and presumed biological properties. To acquire a comprehensive understanding of the biological functions of the matrisome and its components, including proteoglycans and glycosaminoglycans (GAGs), in GBM tumorigenesis, and to identify potential biomarker candidates, we studied the alterations of GAGs, including heparan sulfate (HS) and chondroitin sulfate (CS), the core proteins of proteoglycans, and other glycosylated matrisomal proteins in GBM subtypes vs. control human brain tissue samples. We scrutinized the proteomics data to acquire in-depth site-specific glycoproteomic profiles of the GBM subtypes that will assist in identifying specific glycosylation changes in GBM. We observed an increase in CS 6-O sulfation and a decrease in HS 6-O sulfation, accompanied by an increase in unsulfated CS and HS disaccharides in GBM vs. control samples. Several core matrisome proteins, including proteoglycans (decorin, biglycan, agrin, prolargin, glypican-1, CSPG4), tenascin, fibronectin, hyaluronan link protein 1 and 2, laminins, and collagens, were differentially regulated in GBM vs. controls.
Interestingly, a higher degree of collagen hydroxyprolination was also observed for GBM vs. controls. Further, two proteoglycans, CSPG4, and agrin were significantly lower, about 6–fold for IDH-mutant, compared to the WT GBM samples. Differential regulation of O-glycopeptides for proteoglycans, including brevican, neurocan, and versican, was observed for GBM subtypes vs. controls. Moreover, an increase in levels of glycosyltransferase and glycosidase enzymes was observed for GBM when compared to control samples. We also report distinct protein, peptide, and glycopeptide features for GBM subtypes comparisons. Taken together, our study informs understanding of the alterations to key matrisomal molecules that occur during GBM development.
Corresponding author- Prof. Joseph Zaia, Dept. of Biochemistry, Center for Biomedical Mass Spectrometry, Boston University.
Collaborator- Prof. Joanna Phillips, Dept. of Neurological Surgery, Brain Tumor Center, Helen Diller Family Cancer Research Center, University of California San Francisco, Division of Neuropathology, Department of Pathology, University of California San Francisco.
Remi Janicot awarded American Heart Association fellowship
Remi Janicot in the Garcia-Marcos laboratory was awarded an American Heart Association (AHA) predoctoral fellowship for his project "Optical biosensor platforms for the direct interrogation of GPCR signaling in cardiovascular cells”
The goal of this project is to create new experimental tools that can transform how GPCRs are studied. The main benefit over current methods is that these tools can be easily used in cell models that are relevant to study heart or lung disease, which has been an important limitation in the field. These new experimental possibilities would open new doors for the entire cardiovascular research community. The tools designed in this project will allow to study GPCRs with fidelity and precision. This would pave the way to develop new drugs to treat life-threatening cardiac disorders.
Congratulations Remi!
Kristen Segars Receives F30 Grant for Work with Diabetic Corneal Epithelia
Congratulations to Kristen Segars, an MD/PhD candidate in Dr. Trinkaus-Randall’s lab, for her receipt of an F30 grant for her work, “Delayed wound healing in diabetic corneal epithelia: reduction in protein response after injury and uncoordinated cell-cell communication."
Non-healing corneal injuries affect up to 70% of patients with type 2 diabetes, representing a significant cause of vision loss in this population. Although there are treatments available to improve the symptoms of poorly-healing corneal wounds, the only permanent solution is a corneal transplant, a procedure not readily available worldwide. By examining changes in various proteins involved in the wound healing response at the cellular level, Segars hopes to understand why the corneas of diabetic patients fail to heal effectively.
When an otherwise healthy cornea is injured, cells next to the wound experience a number of changes necessary for coordinated migration and wound closure. One change involves the activation of the purinergic receptor, P2X7, found on the cell surface. When active, P2X7 generates a specific pattern of calcium signaling events that travel from cell to cell through activation of the ion channel Pannexin-1. These propagated signaling events represent cell-cell communication, and ultimately lead to re-arrangement of cytoskeletal proteins and coordinated wound closure. Previous studies have identified aberrant localization and activation of P2X7 in pre-type 2 diabetic models. We have preliminary evidence that the signaling profile of wounded diabetic cells lacks the characteristic P2X7 signaling response. This was confirmed using specific agonists to P2X7, and observing a greatly diminished response in diabetic cells. The goal of this proposal is to uncover how the cell-cell communication events in the P2X7 signaling cascade are regulated, how this regulation is thrown off in diabetic systems, and how this change in regulation affects actin bundling and ultimately cell motility.
Segar's preliminary data has identified a set of cells that she speculates are controllers or leader cells, as they initiate communication events in neighboring cells, and propagated signaling events are greatly reduced in their absence. Aim 1 will use a machine learning approach to investigate the presence of these leader cells in both diabetic cell culture and corneal models. Aim 1 will also address the role of Pannexin-1 in the generation of a unique leader cell signaling profile. Furthermore, the downstream impact of P2X7/Pannexin-1 signaling will be assessed by using 3D electron microscopy to study actin arrangement in wounded diabetic and control corneas. In Aim 2, the localization of P2X7 and Pannexin-1 protein and mRNA within the cells of corneal samples will be examined. This will yield data regarding both general trends in expression between diabetic and control groups, and differences in expression within a single sample that may explain the functional difference between leader cells and the rest of the epithelial sheet. In addition, Aim 2 will address whether the co-localization of P2X7 and Pannexin-1 proteins (before and after a wound) is necessary for wound repair. Together these Aims will produce significant advances in our understanding of the regulation of the P2X7/Pannexin-1 signaling cascade, alterations at the mRNA, protein, and functional level of this cascade in diabetes, and downstream effects of these aberrations on the actin cytoskeleton.
Obesity-induced senescent macrophages and fibrosis
As organisms grow, older cells can undergo a phenomenon called senescence. This process defines a cell state where cells permanently stop dividing but do not die. Senescent cells secrete toxic pro-inflammatory factors contributing to the development of many diseases.
BUSM researchers have shown that obesity in experimental models led to senescence of macrophages, an immune cell subtype within fat or adipose tissue published online in the journal Life Science Alliance. Obesity-induced senescent macrophages activate a fibrotic transcriptional program in adipocyte progenitors by Nabil Rabhi, Kathleen Desevin, Anna C Belkina, Andrew Tilston-Lunel, Xaralabos Varelas, Matthew D Layne, Stephen R Farmer
According to the researchers, the fact that macrophages can become senescent is an unexpected finding. Many of the macrophages within obese tissue were senescent and those senescent cells may be a significant driver of fat tissue fibrosis. These findings suggest that obesity accelerates cellular or biological immune aging in fat.
“In healthy individuals, those cells contribute to cleaning the tissue from dead adipocytes (cells specialized for the storage of fat) and help in the cellular turnover. We demonstrated that macrophages lost this capacity when they become senescent,” explained first and co-corresponding author Nabil Rabhi, PhD, an instructor of biochemistry at BUSM.
The researchers also found that senescent macrophages secrete a variety of factors, one of which is a molecule called osteopontin which they found is responsible for adipose tissue fibrosis. “Our finding suggests that macrophages ages faster in obese animals. This accelerated senescence may contribute to the pathological thickness or fibrosis of fat tissue observed in obese individuals with type 2 diabetes,” said Rabhi.
The researchers believe understanding new regulatory pathways that control adipose tissue responses to obesity may help identify new targets for obesity treatment. “Our finding indicates that targeting the senescent macrophages population or using osteopontin inhibition may represent a promising approach for obesity treatment and its adverse complication including type 2 diabetes,” added Rabhi.
Dmitry Kretov selected for early career development award
Congratulations to Dmitry Kretov for being selected for a Department of Biochemistry Early Career Development Award. Dmitry is an Instructor working in the Cifuentes laboratory on a project entitled: “Development of an integrative approach to reveal the specificity of RNA-binding proteins and their effect on mRNA stability and translation in vivo”. This award is $21,000 for 1 year.