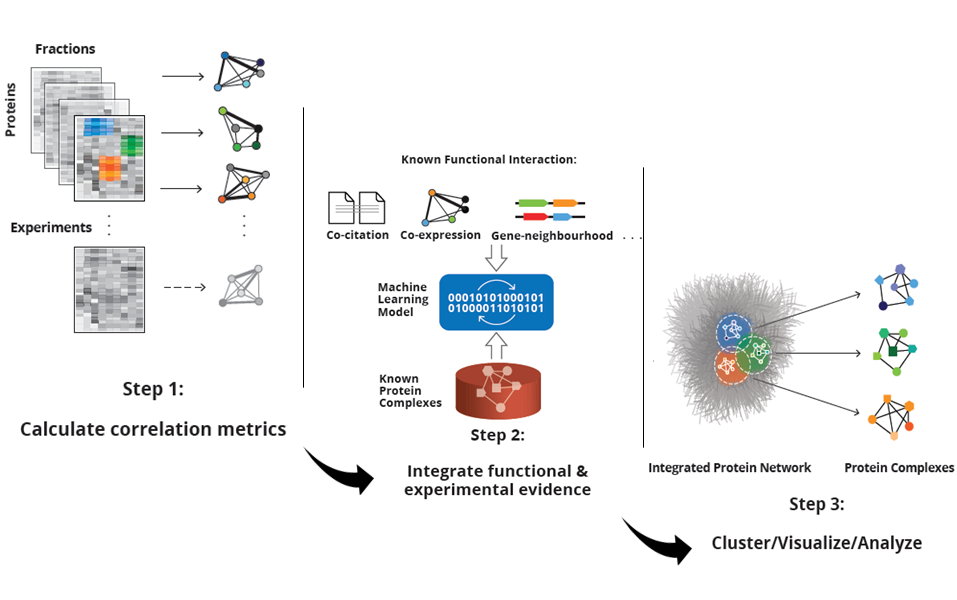
A recent publication in Nature Methods by Hu et al. from the Emili lab describes a software toolkit that enables users to infer protein complexes from elution profile-based fractionation methods. Over the last couple of years methods to identify macromolecular complexes without antibody pulldowns or affinity-purification have been described that are based on the idea to fractionate cellular protein extracts under non-denaturing conditions and to quantify the abundance profiles of the proteins across the fractions. Co-fractionating proteins are then considered to be members of the same protein complex. Inferring protein complexes from co-fractionation data is a non-trivial computational task, however. The software (‘EPIC’) helps to automate this process and simplify the discovery of protein complexes. This study describes the computational methods and an application to C. elegans in order to reveal a map of putative worm protein complexes. The novelty here is both the specific software toolkit, which is freely available online, and the global worm interactome.