Mosquitos and Small RNAs
BUSM researchers are now able to provide a new genomics resource that details the small RNA transcriptomes (gene expression) of four bio-medically important mosquito species.
This is the first study to provide a platform for biologists to compare the characteristics of these small RNAs between these four mosquitoes as well as the most widely used insects for genetic experiments, the fruit fly, Drosophila. Although previous studies looked at each of the individual mosquito species separately, this study is the first to allow comparisons between all four species.
“Although mosquitoes are related to Drosophila, they have very different genomes. In addition, mosquitos bite humans for blood meals that allow them to reproduce and but unfortunately allows serious human pathogens like viruses to infect us and cause diseases like yellow fever virus, dengue fever virus, zika virus and eastern equine encephalitis virus,” explained corresponding author Nelson Lau, PhD, associate professor of biochemistry.
The researchers obtained cell cultures and dissected samples of the mosquito species Anopheles gambiae, Culex quinquefasciatus, Aedes aegypti and Aedes albopictus. They extracted and purified the small RNA molecules, created libraries for high-throughput sequencing, and then developed a special bioinformatics platform to provide thorough genomic analysis of these small RNAs. They provide all this analysis in a public database website .
The four mosquito species have global impacts on human health. Anopheles is the major vector for the parasite causing malaria, but is not known to transmit many viruses. In contrast, Culex and Aedes mosquitoes are well known to pass viruses between humans during mosquito bites, but it is still unknown why there is this difference between mosquito species for this capacity to spread viruses.
According to the researchers this study will allow for better biochemical studies in mosquito cells. “If we can find weaknesses in the small RNA pathways of mosquitoes to make them more intolerant of viruses, perhaps they won’t be so able to pass the virus from biting one human to the next human victim.”
This study was a collaboration between the Lau lab in the Department of Biochemistry and the John Connor and Tonya Colpitts labs of the BU National Emerging Infectious Disease Laboratory (NEIDL) as well as many other mosquito biologists in the USA and the United Kingdom.
The findings appear online in the journal Genome Research.
Dr. Barbara Schreiber—Distinguished Faculty of the Month
The Faculty Affairs Office announced that Associate Professor of Biochemistry Barbara M. Schreiber, PhD, is September’s Distinguished Faculty of the Month.
Dr. Schreiber came to BUSM in 1975 as a graduate student in Microbiology and earned her PhD in Microbiology in 1981. She was a post-doctoral trainee in Clinical Microbiology at University Hospital from 1981-83 and a postdoctoral fellow in Biochemistry from 1983-87. Dr. Schreiber was appointed research assistant professor in 1987, research associate professor in 1997 and associate professor in 1999.
Dr. Schreiber currently serves as Director of Biochemistry graduate studies, Director of the Graduate Medical Sciences (GMS) PhD Program in Biomedical Sciences (PiBS), and course manager of the biochemistry class taught to first-year BU dental students and Oral Health Sciences MS students. Dr. Schreiber is the PI on a Burroughs Wellcome Fund Career Guidance in Training award and was co-PI on the NIH-funded BU’s BEST award, dedicated to broadening experiences in biomedical science training for our PhD students and post-doctoral trainees. She also serves as Assistant Dean for GMS Alumni Affairs.
The nomination states, “Dr. Schreiber orchestrated voluntary, weekly video conferences/meet-ups among the first-year Biomedical PhD (PiBS) students that we all found to be engaging and intellectually stimulating throughout the COVID-19 adjustment period.”
“Dr. Schreiber provided us with some personal interaction during a time of government mandated social isolation and general ambiguity from the earliest stages of the pandemic.”
Congratulations!
See original post: https://www.bumc.bu.edu/busm/2020/08/31/dr-barbara-schreiber-awarded-september-distinguished-faculty-of-the-month/
Research discoveries: DOT in worm chromatin
A new study from the Grishok lab: “Caenorhabditis elegans Deficient in DOT-1.1 Exhibit Increases in H3K9me2 at Enhancer and Certain RNAi-Regulated Regions” by Ruben Esse and Alla Grishok has recently been published in a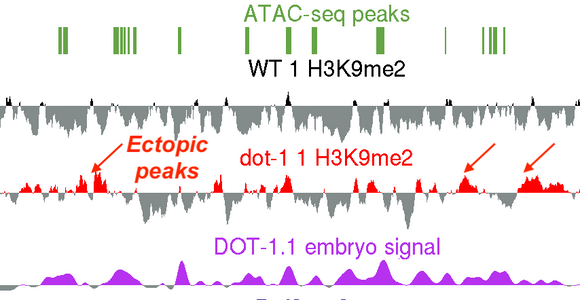 Special issue of journal Cells (MDPI) “Heterochromatin Formation and Function”. The lab has previously shown that Histone H3 lysine 79 methyltransferase DOT-1.1 (an orthologue of human DOT1L) is localized to developmental enhancers in nematodes and promotes lineage-specific, most notably neural, gene expression. The new report describes ectopic deposition of a heterochromatin mark at the enhancers in the absence of DOT-1.1 and H3K79 methylation and suggests that H3K79 methylation “poises” developmental genes for future activation by lineage-specific transcription factors. Notably, the known role of DOT1L in promoting HOXA gene expression (both in normal and cancer contexts) fits the general model of developmental gene activation by DOT1L suggested by the new study by Esse and Grishok.
Special issue of journal Cells (MDPI) “Heterochromatin Formation and Function”. The lab has previously shown that Histone H3 lysine 79 methyltransferase DOT-1.1 (an orthologue of human DOT1L) is localized to developmental enhancers in nematodes and promotes lineage-specific, most notably neural, gene expression. The new report describes ectopic deposition of a heterochromatin mark at the enhancers in the absence of DOT-1.1 and H3K79 methylation and suggests that H3K79 methylation “poises” developmental genes for future activation by lineage-specific transcription factors. Notably, the known role of DOT1L in promoting HOXA gene expression (both in normal and cancer contexts) fits the general model of developmental gene activation by DOT1L suggested by the new study by Esse and Grishok.
Welcome New Faculty—Michael Blower
We are delighted to announce the recruitment of Michael Blower, PhD who will be joining the Department in September 2020 as an Associate Professor. Dr. Blower was recruited through a joint search of the Biochemistry Department and the Genome Science Institute.
The long term goal of the Blower lab is to understand how cell division changes gene expression in mitotic and meiotic cells, and how the process of transcription affects chromosome segregation. They are currently focused on: 1) how transcription is silenced during mitosis and restarted following mitosis, 2) how non-coding RNAs are removed from chromatin during mitosis, 3) regulation of gene expression in oocytes by cytoplasmic polyadenylation.
They are a multidisciplinary lab that uses cell biology, biochemistry, and genomic approaches coupled with CRISPR genome engineering. The Blower lab will be located on the first floor of the K (Conte) building.
Please join us in welcoming Mike and his lab to the department!
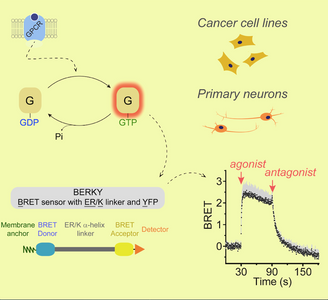
Research discoveries: seeing G-protein signaling
COVID-19 Research
Biochemistry labs are actively engaged in COVID-19 related research.
The Emili laboratory is using systems biology and quantitative mass spectrometry-based approaches to study how the virus hijacks the host cellular protein machinery via a network of viral protein interactions with human cell surface receptors as well as intracellular signaling, metabolic and biomolecular replicative pathways, and so find actionable targets to boost adaptive cell- and tissue-level host responses in human and animal models. The are also using chemical proteomics to characterize potential anti-viral ligands and study the mechanism-of-action of all bioactive compound leads or ‘hits’ emerging from ongoing screens by our collaborators at the NEIDL to enhance their translational impact.
The Saeed laboratory is screening a library of protease inhibitors originally developed against other viruses to test their ability to inhibit SARS-CoV-2. They are taking several molecular virology approaches to understand which lung cells are preferentially targeted by the virus and what molecular mechanisms underlie the disease pathogenesis.
The Lau lab is also collaborating with the Saeed lab to build new human cell lines with fluorescent reporter genes to be inserted into the endogenous locus of several Interferon Stimulated Genes (ISGs). These ISGs are critical response genes triggered by cells responding to an acute viral infection and interferon signaling, but a proper subsequent biological response is for cells to turn back down the expression of ISGs after the first reaction to the virus. When cells or human patients are unable to turn off activated ISGs, this severe “cytokine storm” effect can be seen in certain severe cases of COVID19. With the engineered human cell lines with tagged ISGs available, they will perform real time monitoring of the cellular response during a lab-controlled COVID19 infection.
How can I help?
If you are interested in supporting these or other research efforts in the Department please contact any of our faculty, or click here for philanthropic opportunities to support our research.
Research discoveries: connective tissue assembly
A new study from the Layne laboratory: "Mechanisms of aortic carboxypeptidase-like protein secretion and identification of an intracellularly retained variant associated with Ehlers–Danlos syndrome" was published in The Journal of Biological Chemistry. This study identified that a defective form of the secreted protein aortic carboxypeptidase-like protein (ACLP) from patients with Ehlers-Danlos syndrome (EDS) is retained in cells and induces cellular stress. This collaborative study was with the Wong and Smith labs from the BU College of Engineering. 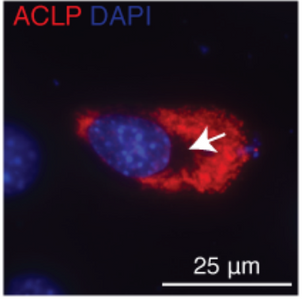
EDS is a prevalent genetic disease that results in weakened connective tissues. This disease results in joint hypermobility, vascular disruption, and aberrant wound healing. Mutations in the gene that encodes for ACLP have recently been identified to cause a novel variant of EDS. Furthermore, excessive amounts of ACLP cause fibrosis in multiple organs including the lung, liver, and adipose tissue.
Detailed characterization of a mutant form of ACLP identified in an EDS patient revealed that this protein was retained in cells. Collagen fibers were generated in vitro that contained ACLP and compared to collagen only controls, they found that fibers with ACLP exhibited increased mechanical properties consistent with a function in maintaining connective tissue integrity. This study was also the first to show that ACLP contributes to the mechanical strength of collagen fibers that make up numerous connective tissues including ligaments, tendons, and cartilage.
Henry I. Russek Achievement Day Awards
The Department of Biochemistry Henry I. Russek Achievement Day Awards Committee is pleased to announce this year’s award winners. Julia Hicks-Berthet is the first prize winner, Nathan Kingston is the second prize winner and Deborah Chang is third prize winner. Our congratulations go out to the winners as well as their advisors! Please read about each of the winners below.
Julia Hicks-Berthet—First Prize
Julia is a student in Dr. Xaralabos (Bob) Varelas’s lab. From her research to her citizenship, Julia is a “winner”. Julia’s dissertation work involves the characterization of pathways involved in lung epithelial cell fate. Integrating in vitro (primary cell models), in vivo (knockout mouse models) and computational (ChIP Seq analysis) approaches, the work has resulted in the identification of a novel role for the Hippo pathway transcriptional effectors Yap and Taz in the regulation of goblet epithelial cells, with implications in asthma, cystic fibrosis and bronchitis. In the future, Julia looks forward to developing therapeutic tools in regenerative medicine. A great citizen, Julia’s participation ranges from mentoring others in the lab to participating as a discussion leader in the Foundations in Biomedical Sciences curriculum, tutoring dental students and serving on the department’s seminar committee. Julia’s passion for science is infectious!
Nathan Kingston—Second Prize
Nathan (Nate) is a student in Dr. Xaralabos (Bob) Varelas’s lab. Nate is studying the role of Taz and Yap, Hippo pathway transcriptional effectors, in idiopathic pulmonary fibrosis. Cell populations that contribute to pulmonary fibrosis are poorly understood. Nate’s studies have identified roles for Taz and Yap in a population of PDGFRbeta-expressing mesenchymal cells that promotes matrix deposition in a model of bleomycin-induced lung injury. His studies have shown that loss of Taz or Yap in these cells protects against fibrosis-induced alveolar damage. Nate looks forward to becoming an independent researcher, either in an academic or industry setting. A great collaborator and citizen, Nate was in leadership positions in the Biomedical PhD Student Organization (BPSO), served as a peer mentor to incoming PhD students, mentored many in the lab and tutored DMD students. Nate’s collaborative nature enables science innovation!
Deborah Chang—Third Prize
Deborah is a student in Dr. Joseph Zaia’s lab. Developing an effective influenza A virus vaccine remains a challenge. With the goal of enabling a path to the development of more effective vaccines, Debbie integrates biochemical, mass spectrometry and bioinformatic approaches to design a method to quantify glycoforms in various strains of the virus to understand how it escapes adaptive immunity. Debbie looks forward to working in a mass spectrometry core facility in an academic setting, collaborating on a variety of research projects. A great citizen, Debbie was in a leadership position in the Biomedical PhD Student Organization (BPSO), she volunteers during PhD recruitment and orientation events, serves as a peer mentor for first year PhD students and for a rotation student in the lab, and she served on the department retreat planning committee. Debbie is intellectually curious and embraces all opportunities to add tools to her armamentarium!
Emili Lab Receives CTSI Covid Pilot Grant
The Boston University Clinical and Translational Science Institute (BU CTSI) has awarded a pilot grant to Andrew Emili in collaboration Cathy Costello's laboratory for COVID-related research. The title of the project is "Defining the Spike-ACE2 Glycoprotein Interface Driving SARS-CoVID-2 Infection"
New research discoveries: A small RNA makes blood
Recent research from the Cifuentes laboratory that describes the functional role of a non-canonical microRNA-processing pathway has been published in Molecular Cell: Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis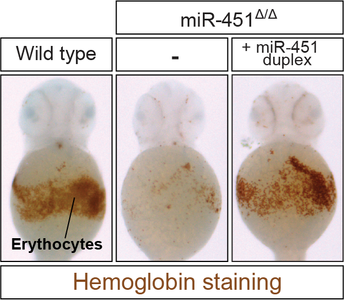
The work spearheaded by the Cifuentes’ laboratory postdoc Dmitry Kretov addresses a long-standing question that has puzzled developmental biologists beyond microRNA aficionados: Why miR-451, a vertebrate-specific microRNA expressed in red blood cells, uses an alternative processing pathway that bypasses Dicer but instead relies on the slicer activity of Ago2?
Kretov et al., report that miR-451 uses this non-canonical processing pathway to escape the global downregulation of microRNAs and Dicer that occurs during erythropoiesis, triggered in part by a negative-feedback loop between miR-144 and Dicer.
Overall, this study uncovers the evolutionary relevance of the non-canonical processing of miR-451 and it highlights a novel physiological role of miR-144, with important clinical implications for the study of anemia and the differentiation of red blood cells.