New research discoveries: Regulators of G protein signaling in cancer
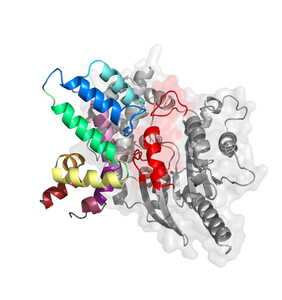 The Garcia-Marcos laboratory has recently published a new study in the journal Science Signaling. In this study, the authors characterized how cancer-associated mutations in a family of negative regulators of G proteins affect the ability of these regulators to modulate G protein activity. Many of these mutations turn out to be deleterious for the G protein regulatory function. In the words of the journal's Editor:
The Garcia-Marcos laboratory has recently published a new study in the journal Science Signaling. In this study, the authors characterized how cancer-associated mutations in a family of negative regulators of G proteins affect the ability of these regulators to modulate G protein activity. Many of these mutations turn out to be deleterious for the G protein regulatory function. In the words of the journal's Editor:
"Mutations in the genes encoding the α subunits of heterotrimeric G proteins are associated with cancer. In particular, mutations that prevent the Gα subunits from hydrolyzing GTP, thus rendering them constitutively active, are pro-oncogenic. DiGiacomo et al. surveyed cancer-associated mutations in regulator of G protein signaling (RGS) proteins, which are physiological inhibitors of G proteins. Through bioinformatics analysis, genetic interaction studies in yeast, and functional assays in mammalian cells, the authors showed that many cancer-associated RGS mutants fail to inhibit G protein signaling because of reduced protein stability or impaired interactions with their targets. With these tools, further cancer-associated mutations in RGS proteins can be characterized."
This work was spearheaded by the former Garcia-Marcos' laboratory postdoc Vincent DiGiacomo, who is currently working in the Cambridge biotech company DeepBiome, with the help of other laboratory members, including two undergraduate students from Boston University. The entire study was carried out in the Department of Biochemistry.
New research discoveries: Yap suppresses tumor T-cells
New research from the Varelas laboratory describes novel roles for the Hippo pathway effector YAP in T cells has been published in PLOS 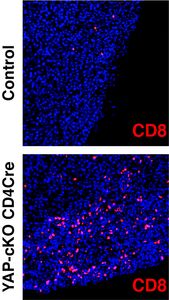 Biology: Yap suppresses T-cell function and infiltration in the tumor microenvironment
Biology: Yap suppresses T-cell function and infiltration in the tumor microenvironment
This study describes previously unappreciated functions for the signaling effector YAP in T cells. Stampouloglou et al., describe immunoinhibitory functions for YAP in CD4+ and CD8+ T cells, which broadly impact T cell functions. T cell specific deletion of the YAP gene was shown to result in the increased activation and differentiation of T cells, which translated in vivo to an increased ability for T cells to block aggressive solid tumor growth. The concept of altering signals within immune cells to boost their ability to fight disease has come to the forefront of medical research, given the effectiveness that many immunotherapies have shown in the clinic. This study places YAP as a key effector of these disease-relevant immune modulating signals. Notably, the authors observed that deletion of YAP resulted in a greatly enhanced ability for T cells to infiltrate solid tumors and become locally activated within the tumor microenvironment. These observations may have important clinical implications, since a major challenge for current immunotherapies is a failure for T cells to effectively infiltrate solid tumors. This study also raises questions about the role of YAP and related signals, such as those mediated by the Hippo signaling pathway, in general T cell biology, opening the door to interesting future research directions.
Infertility in Fruit Flies
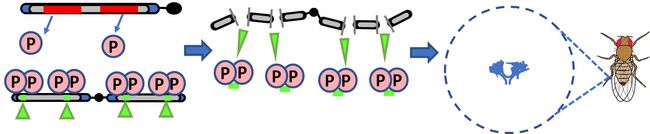 Researchers in the Lau laboratory have discovered a novel parasitic gene in fruit flies that is responsible for destroying the eggs in the ovaries of their daughters. This work is published in eLife.
Researchers in the Lau laboratory have discovered a novel parasitic gene in fruit flies that is responsible for destroying the eggs in the ovaries of their daughters. This work is published in eLife.
Just like fruit flies, human genomes are filled with mobile parasitic genes called transposons and similar to fruit flies, humans use small RNA molecules to silence these genetic parasites so that they can generate proper germ cells for reproduction.
The researchers focused on one parent fly that originated from Harwich, Mass., with the mobile parasitic gene called the P-element. They then generated hybrid offspring between the Harwich fly and a “clean” fly called ISO1 to determine which offspring still caused the infertility syndrome in their daughters and which did not.
They then analyzed the DNA genomes between these two different hybrids and found that Harwich fathers and the sons that still cause infertility in their daughters all had a special hyper mobile version of the P-element that they named the Har-P. “Our discovery of the Har-P element showed that it moves around so extensively in fly germ cells that it causes catastrophic ovary collapse,” explained corresponding author Nelson Lau, PhD, Associate Professor of Biochemistry at Boston University School of Medicine (BUSM).
According to the researchers, human infertility from the incompatibility of two different genomes from the mother and father could be modeled by the infertility syndrome of the Harwich fly fathers mating with ISO1 mothers to cause all their daughters to be infertile. “More than 45 percent of the human genome is made up of remnants of transposons and most of them are properly silenced, but there are still a few active transposons that can move each time a new human is conceived, changing our genomes in a way that is completely different from the general mixing of our fathers and mothers genes during the process of meiosis, when sperm and egg are generated.”
By studying the simpler system of fruit flies where genetic manipulations are easier, the researchers hope to achieve a better understanding of how human genomes are shaped by the multitude of transposons lurking in our genomes and the small RNA molecules we depend upon to keep the transposons in check. They also hope to harness the hyper mobile Har-P element to turn it into a new tool for genetically marking animal cells for developmental biology studies.
Funding for this study was provided by the National Institutes of Health’s Institutes of Aging and Child Health and Human Development .
Building a better flu vaccine
New research from the Zaia laboratory has been published in Molecular Proteomics: "Why glycosylation matters in building a better flu vaccine".
Low vaccine efficacy against seasonal influenza A virus (IAV) stems from the ability of the virus to evade existing immunity while maintaining fitness. While most potent neutralizing antibodies bind antigenic sites on the globular head domain of the IAV envelope glycoprotein hemagglutinin (HA), the error-prone IAV polymerase enables rapid evolution of key antigenic sites, resulting in immune escape. Significantly, the appearance of new N-glycosylation consensus sequences (sequons, NXT/NXS, rarely NXC) on the HA globular domain occurs among the more prevalent mutations as an IAV strain undergoes antigenic drift. The appearance of new glycosylation shields underlying amino acid residues from antibody contact, tunes receptor specificity, and balances receptor avidity with virion escape, all of which help maintain viral propagation through seasonal mutations. The determination of site-specific glycosylation of IAV glycoproteins would enable development of vaccines that take advantage of glycosylation-dependent mechanisms whereby virus glycoproteins are processed by antigen presenting cells
Emili lab—”EPIC” discovery
A recent publication in Nature Methods by Hu et al. from the Emili lab describes a software toolkit that enables users to infer protein complexes from elution profile-based fractionation methods. Over the last couple of years methods to identify macromolecular complexes without antibody pulldowns or affinity-purification have been described that are based on the idea to fractionate cellular protein extracts under non-denaturing conditions and to quantify the abundance profiles of the proteins across the fractions. Co-fractionating proteins are then considered to be members of the same protein complex. Inferring protein complexes from co-fractionation data is a non-trivial computational task, however. The software (‘EPIC’) helps to automate this process and simplify the discovery of protein complexes. This study describes the computational methods and an application to C. elegans in order to reveal a map of putative worm protein complexes. The novelty here is both the specific software toolkit, which is freely available online, and the global worm interactome.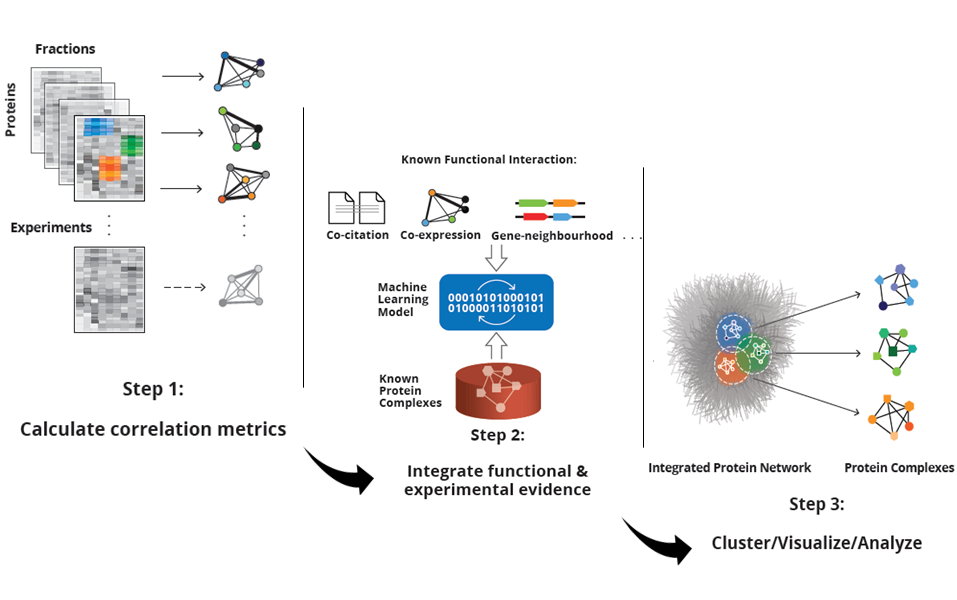
Research Discoveries: VEGFR2 glycosylation
Vascular endothelial growth factor receptor 2 (VEGFR2) is a heavily N-glycosylated pro-angiogenic receptor tyrosine kinase. Stimulation of the receptor by vascular endothelial growth factor (VEGF) induces quiescent endothelial cells to proliferate and sprout, and VEGFR2 signaling is also required for tumor growth and metastasis. In a new paper in the Journal of Biological Chemistry by Kevin Brown Chandler and colleagues in the Costello and Rahimi laboratories demonstrated that the glycosylation status of VEGFR2 alters signaling through the receptor. Specifically, sialic acid-capped N-glycans at site N247 oppose ligand-mediated receptor activation, whereas asialo-glycans (lacking sialic acid) favor VEGFR2 activation.
Welcome New Faculty
The Department wanted to share the good news that Shawn Lyons, Ph.D., has accepted our offer to join the department as an Assistant Professor as of October 1, 2019. Shawn obtained his Ph.D. in the laboratory of Dr. William Marzluff working on histone mRNA metabolism. He is currently a postdoctoral fellow with Dr. Paul Anderson at Harvard Medical School (Brigham and Women’s) working on the regulation of protein synthesis during acute stress response in cells, in particular how RNA metabolism and ribosome biogenesis are altered in response to stress. His work is extremely exciting, and will connect with that of a number of other department members, particularly those in the RNA biology group. Please join us in welcoming Shawn to the department!
Metabolic Regulation Symposium Honoring Paul Pilch— June 24th
Congratulations graduates

Congratulations to the graduates from the Department of Biochemistry. Jessica Calloway Jones from the Farmer lab received her PhD. Neya Vishwanath who performed her research in the Layne lab received her MS in Medical Sciences degree. Neya (picture right) was also a speaker at the morning GMS graduation. Congratulations to all.
Research Discoveries: New ways of activating G-proteins during embryonic development
Heterotrimeric G proteins are signaling switches that control cellular communication 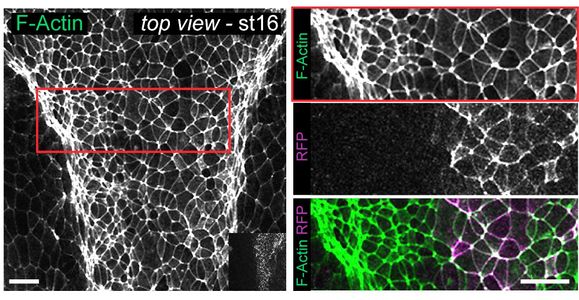 across metazoans. From a traditional standpoint, these G-proteins are activated by G-protein-coupled receptors (GPCRs). However, a recent paper published in the Journal of Cell Biology by Arthur Marivin and colleagues provides direct evidence that heterotrimeric G-proteins can be activated in vivo by a cytoplasmic factor instead of by a GPCR. Specifically, DAPLE, a non-receptor protein bearing an evolutionarily conserved G-protein activating motif, triggers apical cell constriction during neurulation in Xenopus and zebrafish embryos via G-protein dependent signaling. This project of the Garcia-Marcos Lab was carried out in collaboration with the Dominguez Lab (Dept. of Medicine) and the Cifuentes Lab (Dept. of Biochemistry) at BU.
across metazoans. From a traditional standpoint, these G-proteins are activated by G-protein-coupled receptors (GPCRs). However, a recent paper published in the Journal of Cell Biology by Arthur Marivin and colleagues provides direct evidence that heterotrimeric G-proteins can be activated in vivo by a cytoplasmic factor instead of by a GPCR. Specifically, DAPLE, a non-receptor protein bearing an evolutionarily conserved G-protein activating motif, triggers apical cell constriction during neurulation in Xenopus and zebrafish embryos via G-protein dependent signaling. This project of the Garcia-Marcos Lab was carried out in collaboration with the Dominguez Lab (Dept. of Medicine) and the Cifuentes Lab (Dept. of Biochemistry) at BU.
