Bioinformatics for glycomics and glycoproteomics
 The Zaia lab studies protein glycosylation, a mechanism whereby glycans (complex sugar molecules) are attached to proteins through a series of biosynthetic enzyme reactions. Because the reactions do not go to completion, the result is a populations of mature protein molecules heterogeneous with respect to glycosylation and diverse with respect to biological activities. Many families of proteins contain domains (known as lectin domains) that bind to glycan substructures (known as epitopes, containing 1-5 monosaccharide units). Glycosylation of a protein strongly influences its binding to other proteins via lectin domains. The lab is studying the mechanisms whereby viruses alter glycosylation of surface glycoproteins in order to escape host immune response. We have generated large datasets measuring the expression of glycans and glycoproteins from virus evolution experiments, using liquid chromatography-mass spectrometry and tandem mass spectrometry.
The Zaia lab studies protein glycosylation, a mechanism whereby glycans (complex sugar molecules) are attached to proteins through a series of biosynthetic enzyme reactions. Because the reactions do not go to completion, the result is a populations of mature protein molecules heterogeneous with respect to glycosylation and diverse with respect to biological activities. Many families of proteins contain domains (known as lectin domains) that bind to glycan substructures (known as epitopes, containing 1-5 monosaccharide units). Glycosylation of a protein strongly influences its binding to other proteins via lectin domains. The lab is studying the mechanisms whereby viruses alter glycosylation of surface glycoproteins in order to escape host immune response. We have generated large datasets measuring the expression of glycans and glycoproteins from virus evolution experiments, using liquid chromatography-mass spectrometry and tandem mass spectrometry.
- Klein, J.; Zaia, J. Relative Retention Time Estimation Improves N-Glycopeptide Identifications By LC-MS/MS. J Proteome Res 2020, 19, 2113-2121. Pubmed Link
- Klein, J. A.; Zaia, J. A Perspective on the Confident Comparison of Glycoprotein Site-Specific Glycosylation in Sample Cohorts. Biochemistry 2020, . Pubmed Link
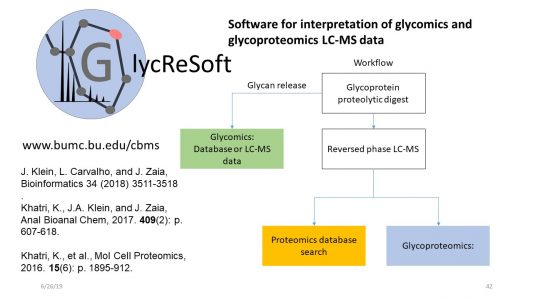
- Klein, J. A.; Zaia, J. Assignment of coronavirus spike protein site-specific glycosylation using GlycReSoft. bioRxiv 2020, , 2020.05.31.125302. Pubmed Link
- Klein, J. A.; Zaia, J. Protocol for analysis of glycoproteomics LC-MS data using GlycReSoft. bioRxiv 2020, , 2020.05.22.108928. Pubmed Link
- Klein, J. A.; Zaia, J. psims – A declarative writer for mzML and mzIdentML for Python. Mol Cell Proteomics 2019, 18, 571-575. Pubmed Link
- Klein, J.; Zaia, J. glypy: An Open Source Glycoinformatics Library. J Proteome Res 2019, 18, 3532-3537. Pubmed Link
- Klein, J. A.; Meng, L.; Zaia, J. Deep sequencing of complex proteoglycans: a novel strategy for high coverage and site-specific identification of glycosaminoglycan-linked peptides. Mol Cell Proteomics 2018, 17, 1578-1590. Pubmed Link
- Klein, J.; Carvalho, L.; Zaia, J. Application of Network Smoothing to Glycan LC-MS Profiling. Bioinformatics 2018, 34, 3511-3518. Pubmed Link
- Hogan, J. D.; Klein, J. A.; Wu, J.; Chopra, P.; Boons, G. J.; Carvalho, L.; Lin, C.; Zaia, J. Software for peak finding and elemental composition assignment for glycosaminoglycan tandem mass spectra. Mol Cell Proteomics 2018, 17, 1448-1456. Pubmed Link
- Khatri, K.; Klein, J. A.; Zaia, J. Use of an informed search space maximizes confidence of site-specific assignment of glycoprotein glycosylation. Anal Bioanal Chem 2017, 409, 607-618. Pubmed Link
- Hu, H.; Khatri, K.; Zaia, J. Algorithms and design strategies towards automated glycoproteomics analysis. Mass Spectrom Rev 2017, 36, 475-498. Pubmed Link
- Zaia, J.; Khatri, K.; Klein, J. A.; Shao, C.; Sheng, Y.; Viner, R. Complete Molecular Weight Profiling of Low Molecular Weight Heparins Using Size Exclusion Chromatography-Ion Suppressor-High Resolution Mass Spectrometry. Anal. Chem. 2016, 88, 10654-10660. Pubmed Link
- Hu, H.; Khatri, K.; Klein, J.; Leymarie, N.; Zaia, J. A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconj J 2016, 33, 285-296. Pubmed Link
- Hu, H.; Huang, Y.; Mao, Y.; Yu, X.; Xu, Y.; Liu, J.; Zong, C.; Boons, G. J.; Lin, C.; Xia, Y.; Zaia, J. A Computational Framework for Heparan Sulfate Sequencing Using High-resolution Tandem Mass Spectra. Molecular & cellular proteomics : MCP 2014, 13, 2490-2502. Pubmed Link
- Maxwell, E.; Tan, Y.; Tan, Y.; Hu, H.; Benson, G.; Aizikov, K.; Conley, S.; Staples, G. O.; Slysz, G. W.; Smith, R. D.; Zaia, J. GlycReSoft: A Software Package for Automated Recognition of Glycans from LC/MS Data. PLoS ONE 2012, 7, e45474. Pubmed Link