Workflow
- Set up a meeting with Drs. Alekseyev and Lenburg to plan your experiment, discuss RNA isolation methods, costs and/or have them order the GeneChips that you need. If you are familiar with microarray analysis, you may want to simply have us order GeneChips for you.
- Isolate either total RNA (most common) or polyA-selected RNA and deliver the sample to Dr. Alekseyev. For examples, see our list of RNA Isolation protocols.
- Wait about 3 days while the microarray resource team labels your RNA, hybridizes it to the GeneChips, collects the data using a laser scanner and presents you with copies of the results.
- Go over basic data analysis with a member of the microarray resource or on your own once you learn how to do it.
RNA Isolation Protocols
Many different RNA isolation protocols are compatible with the Affymetrix GeneChip and custom array sample preparation protocol. The only requirement is that you are able to isolate 50-500 nanograms of high-quality intact RNA that does not contain significant amounts of contaminating DNA.
If you currently have a preferred method for isolating RNA, continue to use it. If you haven’t isolated RNA from your system before and your organism is not on the following list, we can help you identify established protocols that have been used for isolating RNA for Northern blotting or RT-PCR from your system. These types of protocols will also yield “microarray-grade” RNA. Always run an agarose gel of your RNA to assess the quality.
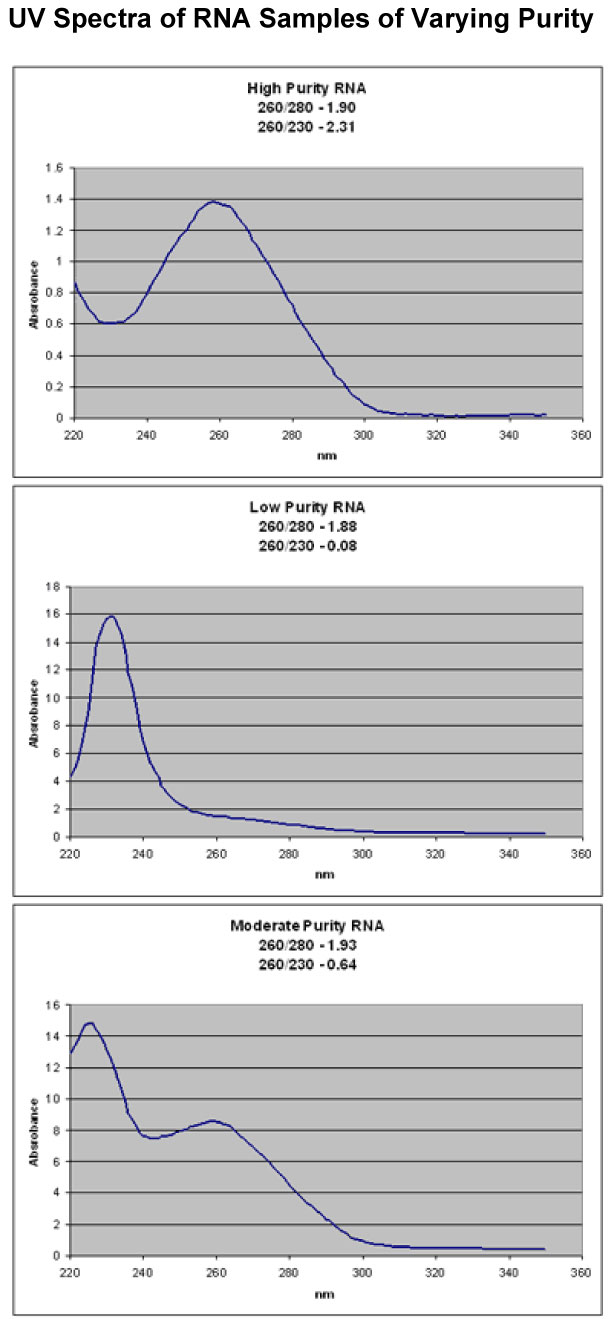
NOTE: If you use a non column based purification reagent such as TRIzol we recommend you precipitate your RNA a second time from ethanol or you pass it through an RNeasy column or similar device. This insures removal of all organic contaminants (See spectra below).
Affymetrix recommends: If going directly from TRIzol-isolated total RNA to cDNA synthesis, it may be beneficial to perform a second cleanup on the total RNA before starting. After the ethanol precipitation step in the TRIzol extraction procedure, perform a cleanup using the QIAGEN RNeasy Mini Kit. Much better yields of labeled cRNA are obtained from the in vitro transcription and labeling reactions when this second cleanup is performed.
RNA Purity
A260 readings and 260/280 ratios are not enough to ensure high purity RNA. You must take a full UV spectrum. Below are 3 example spectra which all have good 260/280 ratios, but have very different purities. You can also use the 260/230 ratio to help determine the amount of organic contamination in your RNA. It is a much more variable number than the 260/280 ratio, but generally, ratios above 1 indicate good purity.
All three samples appeared clean when just taking the A260 and 260/280 ratios. It only becomes clear that they have very different purities when a full UV spectrum is taken.
Yeast Total RNA
Good quality total RNA from yeast cells can be obtained using the hot phenol protocol described by Schmitt, et al. (1990) Nucl Acids Res, 18:3091-3092. “Hot phenol purification of RNA from yeast:”
- Harvest 10 ml of culture by centrifugation and resuspend in 400 ul of AE buffer (AE buffer : 50 mM Na-acetate, pH 5.3, 10 mM EDTA). Transfer to 1.5ml microfuge tube (use a tube with a screw-on cap).
- Add 40 ul of 10% SDS, vortex. Add equal volume (440 ul) phenol (Schmitt et al. say phenol should be pre-equilibrated with AE buffer but it works fine with regular Tris pH 8-equilibrated phenol).
- Vortex, incubate at 65°C for 4 min. Chill rapidly in a dry ice / ethanol bath until phenol crystals appear, centrifuge for 2 min at maximum speed.
- Transfer upper aqueous phase to fresh microfuge tube, extract with phenol / chloroform at RT for 5 min. To extracted aqueous phase add 3 M Na-acetate, pH 5.3, to final conc. of 0.3 M (i.e., add 40 ul) and 2.5 volumes of ethanol to precipitate the RNA. Wash the pellet with 80% ethanol, dry and resuspend in 20 ul of sterile water. Store at -70°.
Mammalian Cell Culture
Affymetrix recommends using QIAGEN’s RNeasy Total RNA Isolation kit.
Mammalian Tissue
Affymetrix recommends using TRIzol Reagent for the initial purification of RNA from mammalian tissue. This is followed by a subsequent sample “clean-up” using QIAGEN’s RNeasy Total RNA Isolation kit (see above). For example, this protocol was developed by Chuck Perou for isolating RNA from tumor specimens (C.M. Perou et al. 1999. PNAS 96: 9212-9217). This protocol may need to be modified for isolating RNA from other specimens.
- The frozen tissue specimen is removed from the freezer and, using a scalpel, small pieces (approximately 50 – 100 milligrams each) are cut off of the larger tissue specimen while it is still frozen. As the small pieces are cut off, they are immediately placed into 10 – 12mls of TRIzol Reagent (Invitrogen) that is contained within a 50ml Screw Cap Tube at RT. For a given tissue sample, upto 1 gram can be placed into this volume of TRIzol, and typically 0.5 – 1 gram of tissue is used per isolation if possible. Note, that as little as 0.2 gram has been successfully used in this volume but it may be better to use 5 – 8mls TRIzol for specimens of this size (the TRIzol Reagent Protocol calls for 1ml TRIzol per 50 – 100mg tissue processed).
- The tissue sample in TRIzol is homogenized using a PowerGen 125 Tissue Homogenizer (Fisher Scientific) starting at 5000 RPM and gradually going up to approximately 20,000 RPM over a period of 30 – 60 seconds at RT. The homogenization is performed until a homogeneous solution is obtained and very few visible tissue pieces can be seen.
- Incubate at RT for 5-10 minutes after homogenization.
- The TRIzol/tissue homogenate is transferred to a 50ml Oak Ridge Centrifuge tube and centrifuged at 12,000g (10,000RPM on SS34) for 5 – 10 minutes at 4°C.
- Remove the upper fat layer by using a Pasteur pipette hooked up to a vacuum flask (the upper fat layer, if present, has a yellowish appearance while the TRIzol homogenate remains red).
NOTE: From here to Step 12, the protocol is exactly as written in the TRIzol Reagent Protocol.
- Add 0.2ml Chloroform per 1ml TRIzol Reagent used in Step 2. Shake tube vigorously for 15-30 seconds by hand and incubate at RT for 5 minutes.
- Spin at 12,000g for 15 minutes at 4°C. Carefully remove the upper aqueous phase, which contains the total RNA, and place this in a new 50ml Oak Ridge Centrifuge tube.
- Precipitate the RNA by adding 0.5ml Isopropyl alcohol per 1ml TRIzol Reagent used in Step 2. Incubate at RT for 10 minutes, then spin at 12,000g for 10 minutes at 4°C.
- Carefully decant the supernatant and wash the pellet once using 75% EtOH by adding 1ml 75% EtOH per 1ml TRIzol used in Step 2. Spin at 7,500g (7500 RPM in SS34) for 5 minutes at 4°C.
- Carefully decant the supernatant and air dry the pellet for 10 – 20 minutes at RT.
- Resuspend the RNA pellet in 200-300 microliter DEPC-DH20 or DEPC-TE, and take A260 if necessary.